presenilin 2
ZFIN 
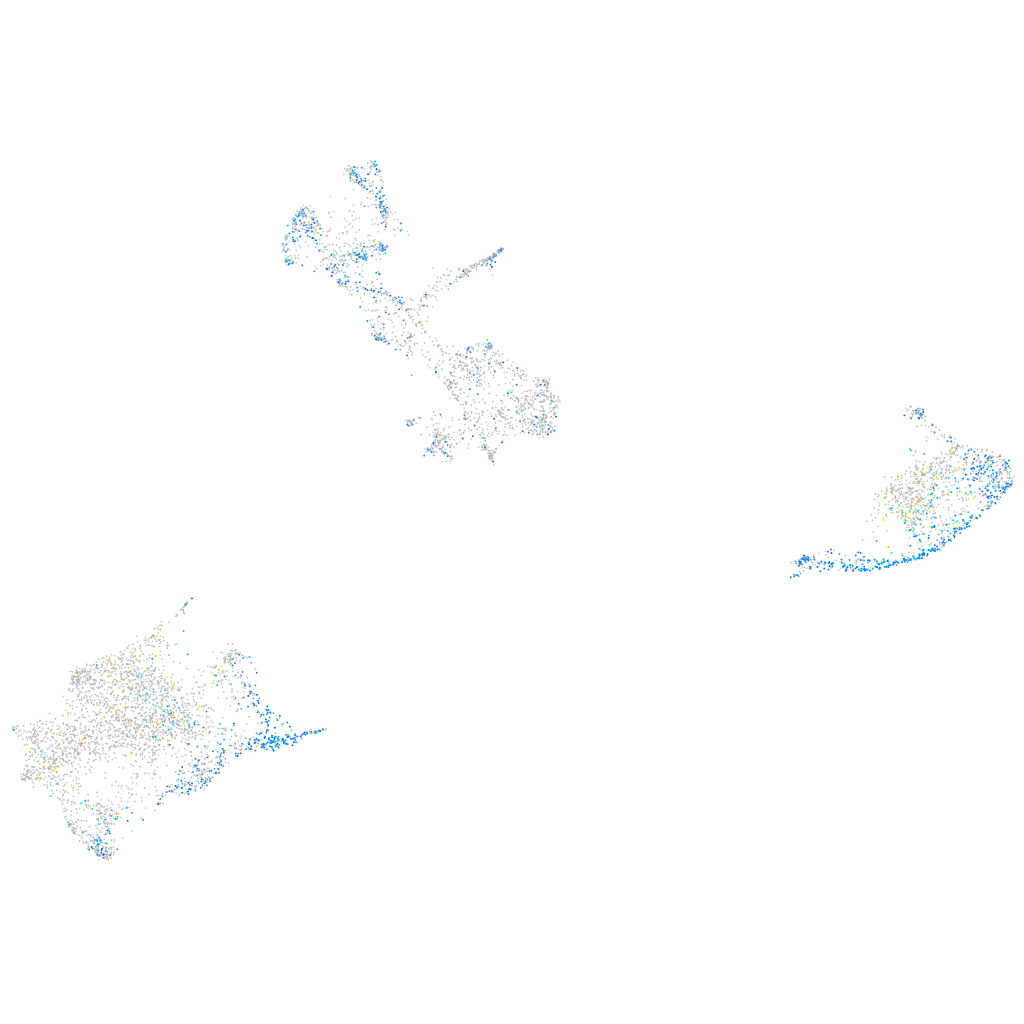
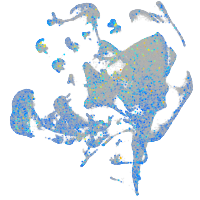
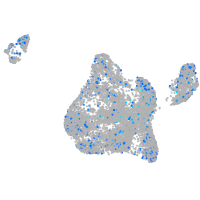
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc45a2 | 0.241 | CABZ01021592.1 | -0.126 |
| tspan36 | 0.236 | si:dkey-251i10.2 | -0.120 |
| tyr | 0.216 | ptmaa | -0.115 |
| slc37a2 | 0.213 | pvalb1 | -0.103 |
| syngr1a | 0.210 | tuba1c | -0.095 |
| psap | 0.207 | hbae3 | -0.095 |
| dct | 0.204 | elavl3 | -0.093 |
| naga | 0.204 | hbbe1.3 | -0.091 |
| atp6ap1b | 0.203 | stmn1b | -0.089 |
| bace2 | 0.202 | nova2 | -0.089 |
| tspan10 | 0.202 | uraha | -0.088 |
| mitfa | 0.201 | aqp3a | -0.086 |
| rab32a | 0.199 | si:ch211-222l21.1 | -0.086 |
| rnaseka | 0.197 | hbbe1.1 | -0.086 |
| msx1b | 0.197 | tmsb | -0.085 |
| pah | 0.196 | hbae1.1 | -0.085 |
| rabl6b | 0.195 | si:ch211-251b21.1 | -0.083 |
| atp6v0ca | 0.195 | pvalb2 | -0.083 |
| atp6v1g1 | 0.194 | gpm6aa | -0.082 |
| ctsba | 0.193 | sncb | -0.080 |
| scpep1 | 0.193 | hmgb1b | -0.078 |
| gpr143 | 0.192 | actc1b | -0.077 |
| pttg1ipb | 0.192 | gng3 | -0.076 |
| slc3a2a | 0.191 | fabp7a | -0.073 |
| degs1 | 0.191 | CU467822.1 | -0.071 |
| pmela | 0.190 | zgc:153031 | -0.071 |
| tyrp1a | 0.189 | epb41a | -0.070 |
| SPAG9 | 0.189 | si:ch73-1a9.3 | -0.070 |
| atp6v1ba | 0.189 | marcksl1a | -0.070 |
| lamp1a | 0.187 | mylz3 | -0.069 |
| rab38 | 0.187 | ndrg2 | -0.069 |
| qdpra | 0.187 | FO082781.1 | -0.067 |
| stx12 | 0.186 | rbfox1 | -0.067 |
| smim29 | 0.184 | cadm3 | -0.066 |
| canx | 0.183 | zc4h2 | -0.065 |