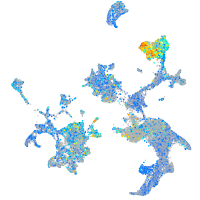
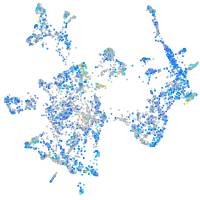
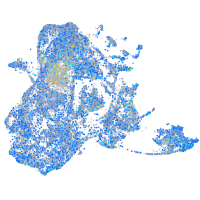
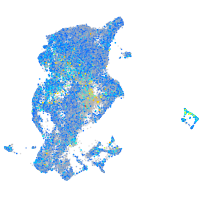
prosaposin
ZFIN 
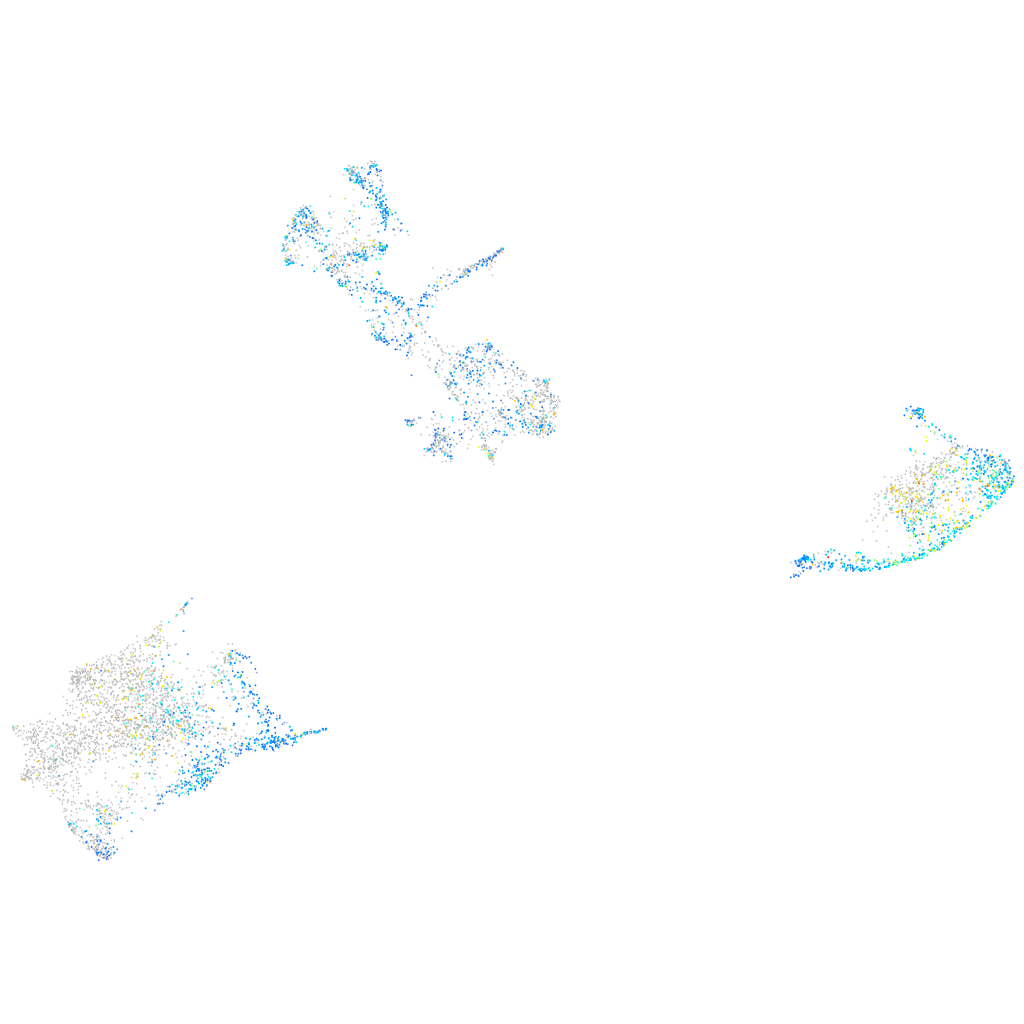
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc45a2 | 0.382 | CABZ01021592.1 | -0.270 |
| tyr | 0.349 | si:dkey-251i10.2 | -0.211 |
| dct | 0.347 | uraha | -0.210 |
| tyrp1a | 0.345 | tmem130 | -0.164 |
| pmela | 0.337 | si:ch211-251b21.1 | -0.154 |
| tspan36 | 0.337 | pvalb2 | -0.153 |
| SPAG9 | 0.336 | pvalb1 | -0.142 |
| slc24a5 | 0.331 | actc1b | -0.142 |
| kita | 0.328 | aldob | -0.141 |
| atp6v0ca | 0.328 | aqp3a | -0.139 |
| slc7a5 | 0.326 | dio3a | -0.135 |
| tyrp1b | 0.324 | hbbe1.1 | -0.131 |
| oca2 | 0.321 | hbae3 | -0.127 |
| slc37a2 | 0.319 | mdh1aa | -0.125 |
| lamp1a | 0.311 | slc22a7a | -0.122 |
| tspan10 | 0.304 | gch2 | -0.120 |
| atp6ap1b | 0.298 | aox5 | -0.119 |
| sypl2b | 0.296 | LOC110437731 | -0.116 |
| aadac | 0.292 | hbbe1.3 | -0.115 |
| slc3a2a | 0.290 | krt18b | -0.113 |
| scpep1 | 0.290 | paics | -0.112 |
| bace2 | 0.288 | zgc:110339 | -0.111 |
| zgc:91968 | 0.286 | hbae1.1 | -0.110 |
| si:ch73-389b16.1 | 0.286 | bco1 | -0.109 |
| ctsba | 0.284 | phyhd1 | -0.108 |
| mtbl | 0.281 | mylpfa | -0.107 |
| zgc:110239 | 0.279 | bscl2l | -0.105 |
| tfap2e | 0.278 | tuba1c | -0.103 |
| pah | 0.275 | zgc:153031 | -0.100 |
| atp6v0a2b | 0.274 | si:dkey-73n8.3 | -0.097 |
| naga | 0.272 | stmn1b | -0.095 |
| rabl6b | 0.272 | tmsb | -0.095 |
| rab38 | 0.272 | plin6 | -0.093 |
| canx | 0.272 | ndrg2 | -0.093 |
| tmem110l | 0.270 | zgc:113337 | -0.092 |