protogenin homolog b (Gallus gallus)
ZFIN 
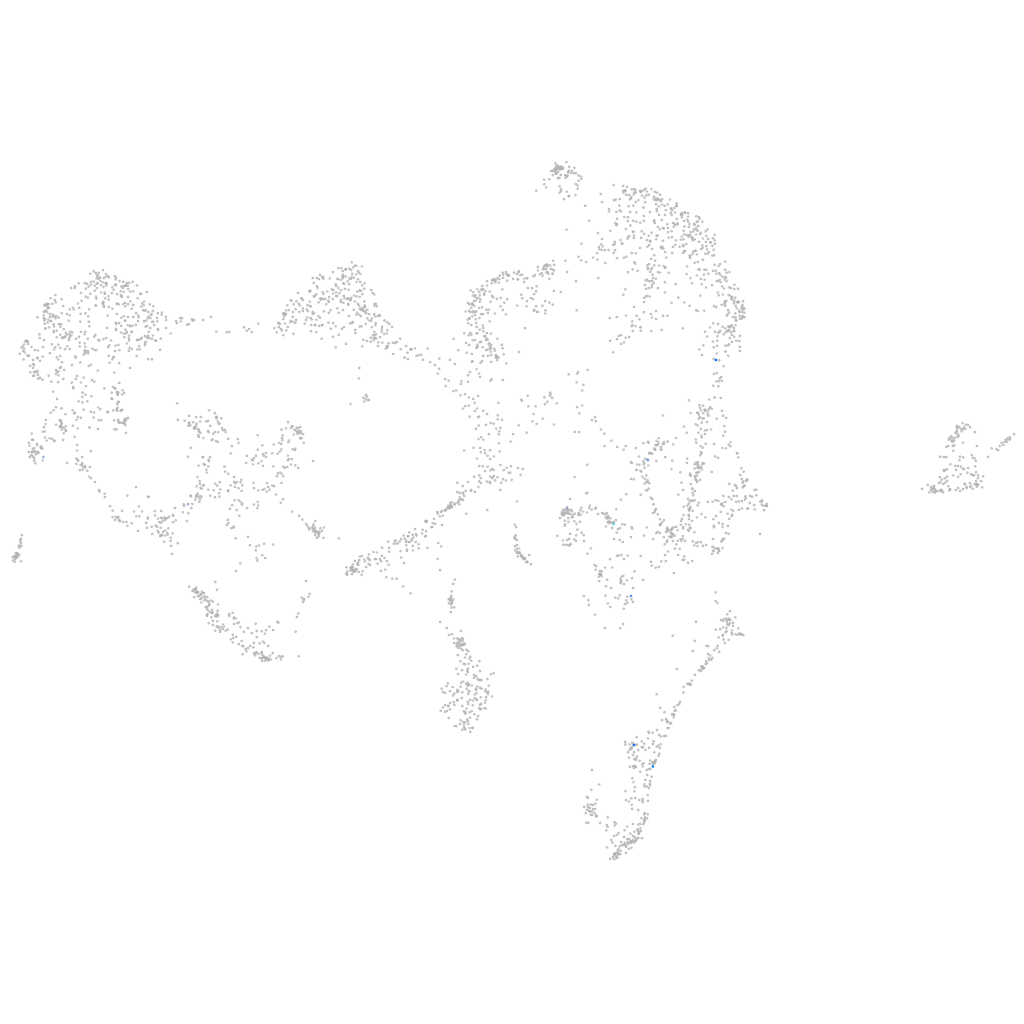
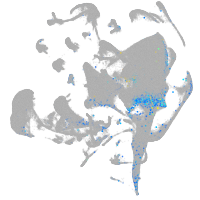
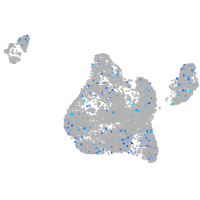
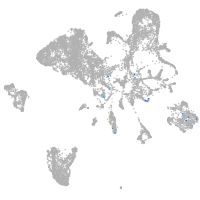
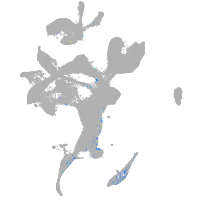
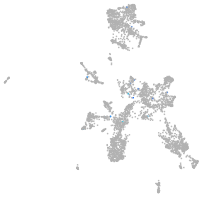
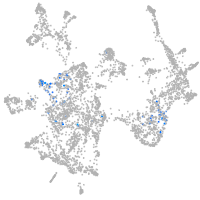
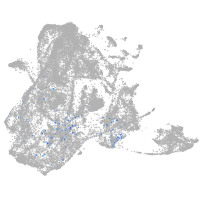
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prdm8 | 0.586 | ldhba | -0.041 |
| BX323559.2 | 0.532 | cycsb | -0.038 |
| prdm13 | 0.498 | cldnb | -0.035 |
| neurog1 | 0.481 | selenow1 | -0.035 |
| gpr17 | 0.456 | gsto2 | -0.033 |
| lbx1b | 0.455 | atp5if1b | -0.033 |
| CU633775.1 | 0.414 | nupr1a | -0.033 |
| CR848812.1 | 0.378 | epcam | -0.033 |
| auts2b | 0.369 | BX000438.2 | -0.032 |
| BX088596.1 | 0.368 | eif4a1b | -0.032 |
| zgc:63568 | 0.366 | ezrb | -0.032 |
| mir219-3 | 0.361 | romo1 | -0.031 |
| LAMP5 | 0.344 | si:ch211-195b11.3 | -0.030 |
| XLOC-000947 | 0.341 | GCA | -0.030 |
| CU468723.1 | 0.336 | prdx5 | -0.030 |
| si:ch211-170d8.2 | 0.335 | oclna | -0.030 |
| sowahca | 0.325 | ak2 | -0.029 |
| sox21a | 0.320 | krt17 | -0.029 |
| si:ch211-57n23.4 | 0.311 | wu:fb18f06 | -0.029 |
| LOC110439327 | 0.288 | TSTD1 | -0.029 |
| jag1a | 0.280 | FQ323156.1 | -0.029 |
| dhx32b | 0.277 | mdh2 | -0.029 |
| pax3b | 0.273 | lxn | -0.029 |
| XLOC-013005 | 0.270 | ndufb7 | -0.028 |
| mapk11 | 0.269 | cd63 | -0.028 |
| pax7b | 0.263 | anxa11a | -0.028 |
| si:ch211-113a14.6 | 0.263 | prelid3b | -0.028 |
| kcnj19a | 0.259 | XLOC-043857 | -0.027 |
| cyp26b1 | 0.257 | ndufaf2 | -0.027 |
| nin | 0.253 | acadm | -0.027 |
| XLOC-006282 | 0.245 | ddt | -0.027 |
| fam117ab | 0.241 | tmx2b | -0.027 |
| LOC101885092 | 0.239 | ugt8 | -0.027 |
| CR751602.2 | 0.238 | cfl1l | -0.027 |
| efcc1 | 0.237 | cbr1l | -0.026 |