phosphoribosyl pyrophosphate synthetase 1A
ZFIN 
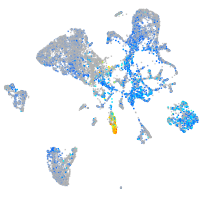
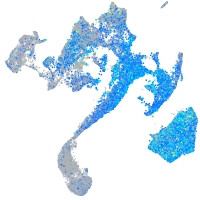
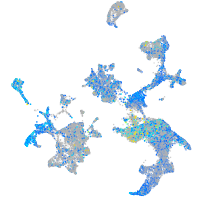
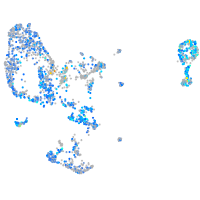
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.352 | CR383676.1 | -0.181 |
| nop58 | 0.309 | gpm6aa | -0.155 |
| snu13b | 0.306 | elavl3 | -0.148 |
| dkc1 | 0.302 | rtn1a | -0.147 |
| nop10 | 0.286 | pvalb1 | -0.144 |
| pcna | 0.279 | myt1b | -0.142 |
| fbl | 0.275 | COX3 | -0.141 |
| nhp2 | 0.271 | pvalb2 | -0.139 |
| hspb1 | 0.269 | actc1b | -0.139 |
| ranbp1 | 0.268 | gpm6ab | -0.133 |
| nop56 | 0.268 | ckbb | -0.127 |
| fen1 | 0.266 | hbbe1.3 | -0.125 |
| cnbpa | 0.261 | stmn1b | -0.124 |
| cdx4 | 0.260 | hbae3 | -0.121 |
| rpa2 | 0.259 | ppdpfb | -0.121 |
| mcm6 | 0.257 | pik3r3b | -0.118 |
| pa2g4a | 0.253 | si:ch211-260e23.9 | -0.118 |
| dut | 0.252 | marcksl1b | -0.115 |
| anp32b | 0.250 | tmsb | -0.114 |
| mcm3 | 0.248 | nova2 | -0.114 |
| mcm5 | 0.248 | elavl4 | -0.113 |
| nutf2l | 0.248 | si:dkeyp-75h12.5 | -0.113 |
| selenoh | 0.247 | mt-co2 | -0.107 |
| mcm7 | 0.247 | tnrc6c1 | -0.106 |
| ncl | 0.246 | mylpfa | -0.105 |
| si:ch211-217k17.7 | 0.245 | tp53inp1 | -0.102 |
| nasp | 0.244 | vim | -0.102 |
| ptges3b | 0.242 | hbae1.1 | -0.102 |
| zgc:56493 | 0.240 | h1f0 | -0.100 |
| cdca7b | 0.239 | scrt2 | -0.099 |
| nop2 | 0.237 | calm1a | -0.099 |
| mcm2 | 0.237 | ndrg2 | -0.099 |
| snrpd1 | 0.236 | hbbe1.1 | -0.098 |
| rpl7l1 | 0.236 | rnasekb | -0.097 |
| mcm4 | 0.235 | gng3 | -0.096 |