protein kinase cGMP-dependent 2
ZFIN 
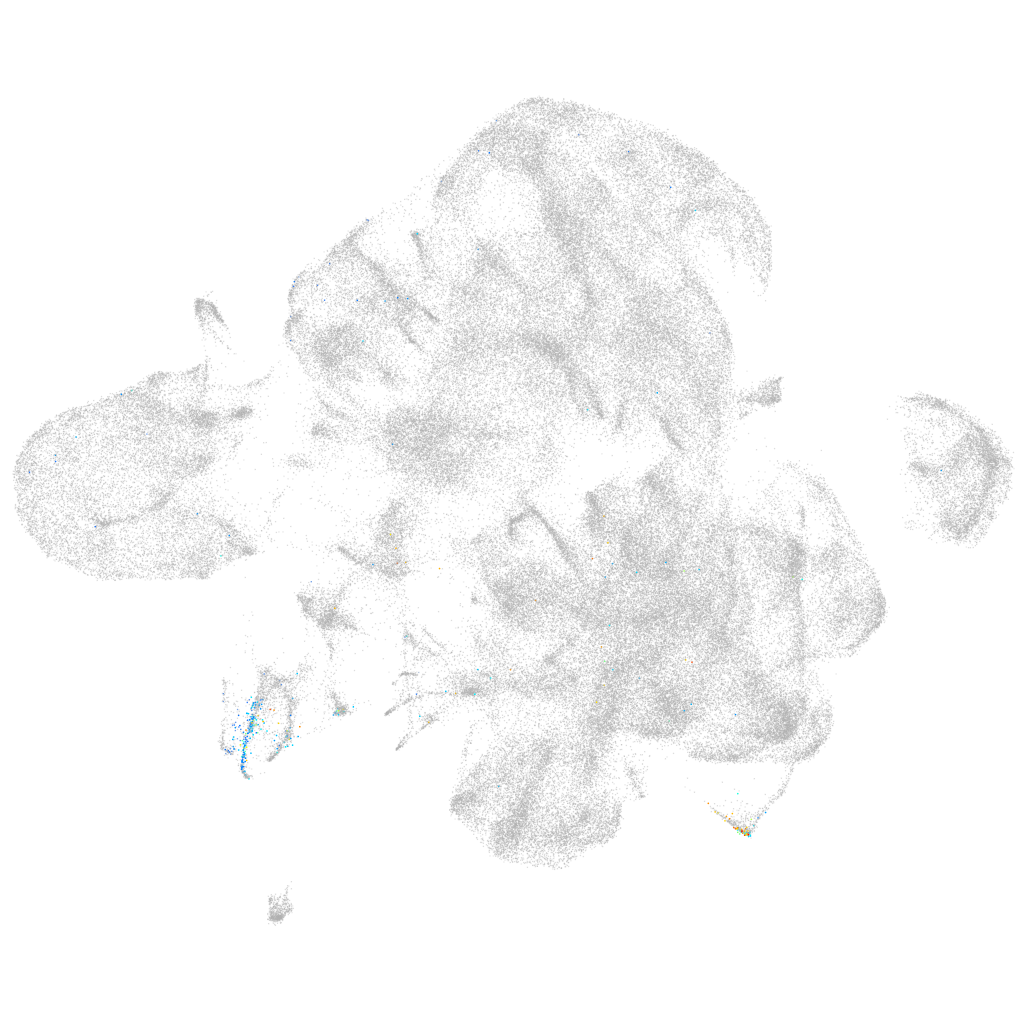
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 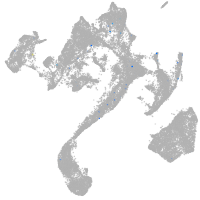 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural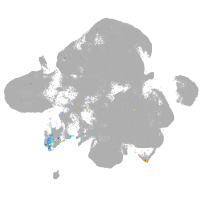 |
spinal cord / glia |
eye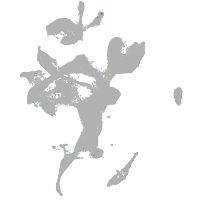 |
taste / olfactory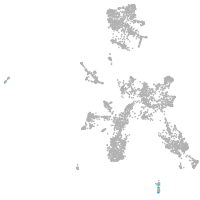 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| g0s2 | 0.192 | si:ch211-222l21.1 | -0.064 |
| gng8 | 0.178 | hmgn2 | -0.042 |
| si:ch1073-90m23.1 | 0.176 | h2afvb | -0.041 |
| eya4 | 0.172 | hmgb2b | -0.038 |
| six2b | 0.156 | hmga1a | -0.036 |
| socs4 | 0.155 | hmgb2a | -0.033 |
| ca4a | 0.149 | cirbpb | -0.033 |
| mmp25b | 0.143 | si:ch211-137a8.4 | -0.033 |
| tac3a | 0.142 | rps12 | -0.032 |
| CABZ01118738.1 | 0.128 | marcksl1a | -0.031 |
| six1b | 0.126 | si:ch73-281n10.2 | -0.031 |
| foxg1d | 0.125 | nova2 | -0.031 |
| slc18a3b | 0.123 | ybx1 | -0.030 |
| CR533578.1 | 0.122 | ran | -0.030 |
| tacr3l | 0.121 | rplp1 | -0.030 |
| cadps2 | 0.116 | hmgb1b | -0.029 |
| cbfb | 0.115 | rpl7 | -0.029 |
| cngb3.1 | 0.114 | rplp0 | -0.029 |
| ngfrb | 0.112 | rps20 | -0.028 |
| GPR151 | 0.110 | cbx3a | -0.028 |
| emb | 0.109 | rps9 | -0.028 |
| ntrk1 | 0.107 | si:ch211-288g17.3 | -0.028 |
| ano2 | 0.105 | cx43.4 | -0.028 |
| kifl | 0.104 | khdrbs1a | -0.027 |
| si:dkeyp-72h1.1 | 0.104 | hmgn6 | -0.027 |
| nwd2 | 0.102 | snrpf | -0.027 |
| sst6 | 0.102 | her15.1 | -0.027 |
| prkg1a | 0.101 | ddx39ab | -0.027 |
| synpr | 0.101 | si:ch73-1a9.3 | -0.027 |
| CABZ01037174.1 | 0.099 | rplp2l | -0.027 |
| prph | 0.099 | rpl32 | -0.027 |
| prkcaa | 0.096 | rps5 | -0.026 |
| si:ch211-56a11.2 | 0.096 | hnrnpabb | -0.026 |
| slc17a7b | 0.095 | rpl11 | -0.025 |
| tspan2b | 0.094 | XLOC-003690 | -0.025 |