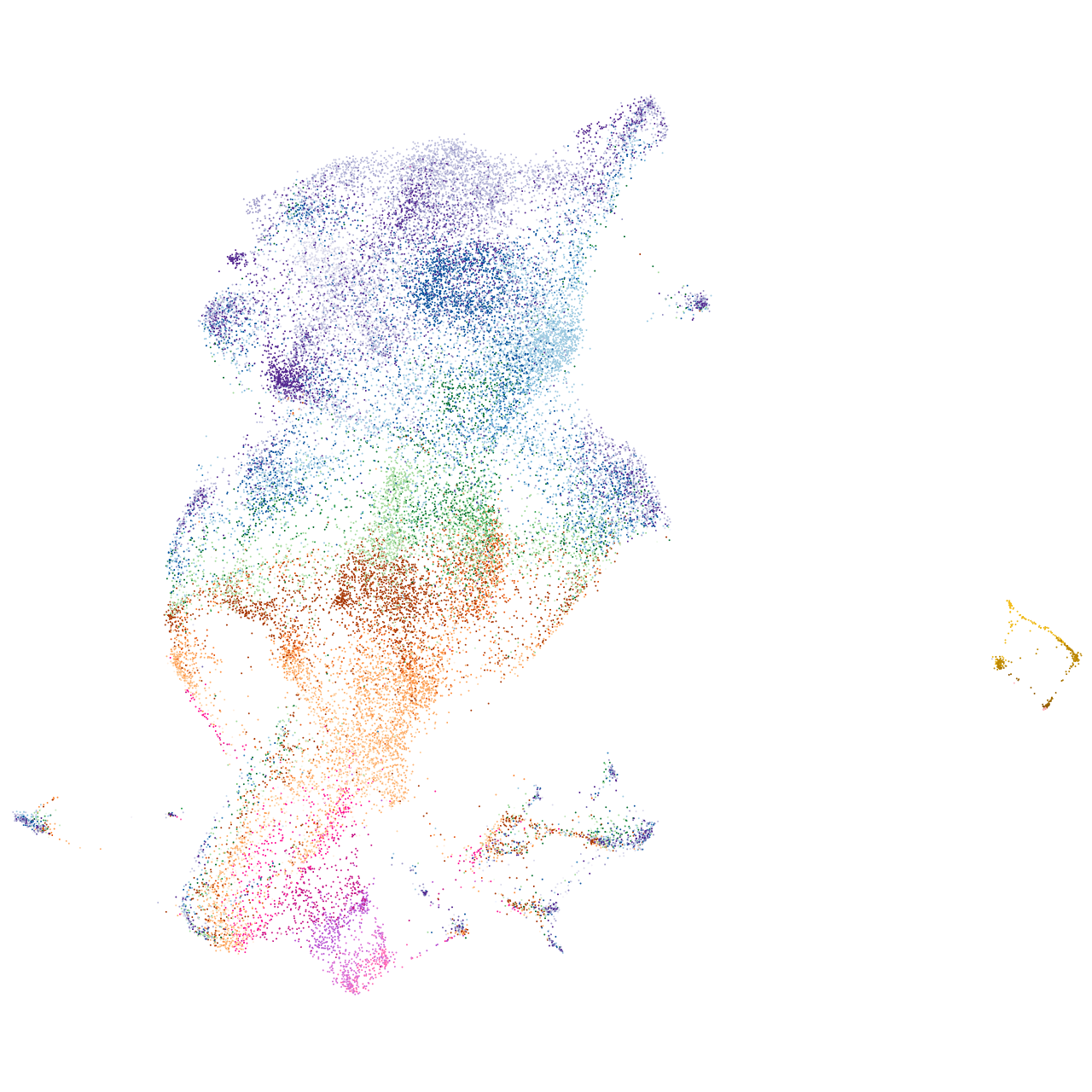"protein phosphatase 6, regulatory subunit 3"
ZFIN 
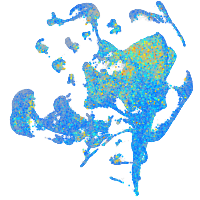
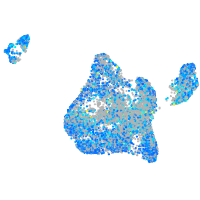
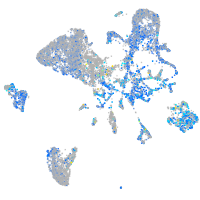
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.151 | cyt1 | -0.121 |
| mt-cyb | 0.148 | krt4 | -0.116 |
| tubb2b | 0.145 | cyt1l | -0.106 |
| mki67 | 0.145 | si:ch211-195b11.3 | -0.098 |
| hmgb2b | 0.141 | krt17 | -0.087 |
| mt-nd1 | 0.139 | rpl39 | -0.087 |
| mt-co2 | 0.138 | krtt1c19e | -0.085 |
| si:ch211-288g17.3 | 0.136 | krt5 | -0.075 |
| mt-atp6 | 0.136 | romo1 | -0.071 |
| tpx2 | 0.133 | icn | -0.071 |
| NC-002333.4 | 0.133 | icn2 | -0.068 |
| plk1 | 0.131 | btf3 | -0.064 |
| aspm | 0.131 | zgc:193505 | -0.064 |
| mt-nd2 | 0.130 | tdh | -0.061 |
| hmgb2a | 0.130 | rpl35 | -0.059 |
| mad2l1 | 0.129 | rplp1 | -0.059 |
| cirbpa | 0.128 | rps29 | -0.058 |
| COX3 | 0.128 | rpl7 | -0.058 |
| cdk1 | 0.128 | ahcy | -0.057 |
| top2a | 0.128 | faua | -0.057 |
| seta | 0.127 | eif4ebp3 | -0.056 |
| ccna2 | 0.125 | si:dkey-16p21.8 | -0.055 |
| kpna2 | 0.124 | agr1 | -0.055 |
| hnrnpaba | 0.123 | krt91 | -0.055 |
| ube2c | 0.123 | rpl22 | -0.055 |
| syncrip | 0.123 | rpl8 | -0.054 |
| lbr | 0.122 | anxa1a | -0.053 |
| birc5a | 0.122 | rpl36a | -0.053 |
| ckap5 | 0.121 | rps26l | -0.052 |
| mt-co1 | 0.121 | rpl37 | -0.051 |
| cenpf | 0.121 | rpl10a | -0.051 |
| kifc1 | 0.120 | eevs | -0.050 |
| ccnb1 | 0.120 | wu:fb18f06 | -0.049 |
| cdca8 | 0.120 | rps2 | -0.049 |
| hmgn2 | 0.119 | si:dkey-151g10.6 | -0.049 |