phosphoribosyl pyrophosphate amidotransferase
ZFIN 
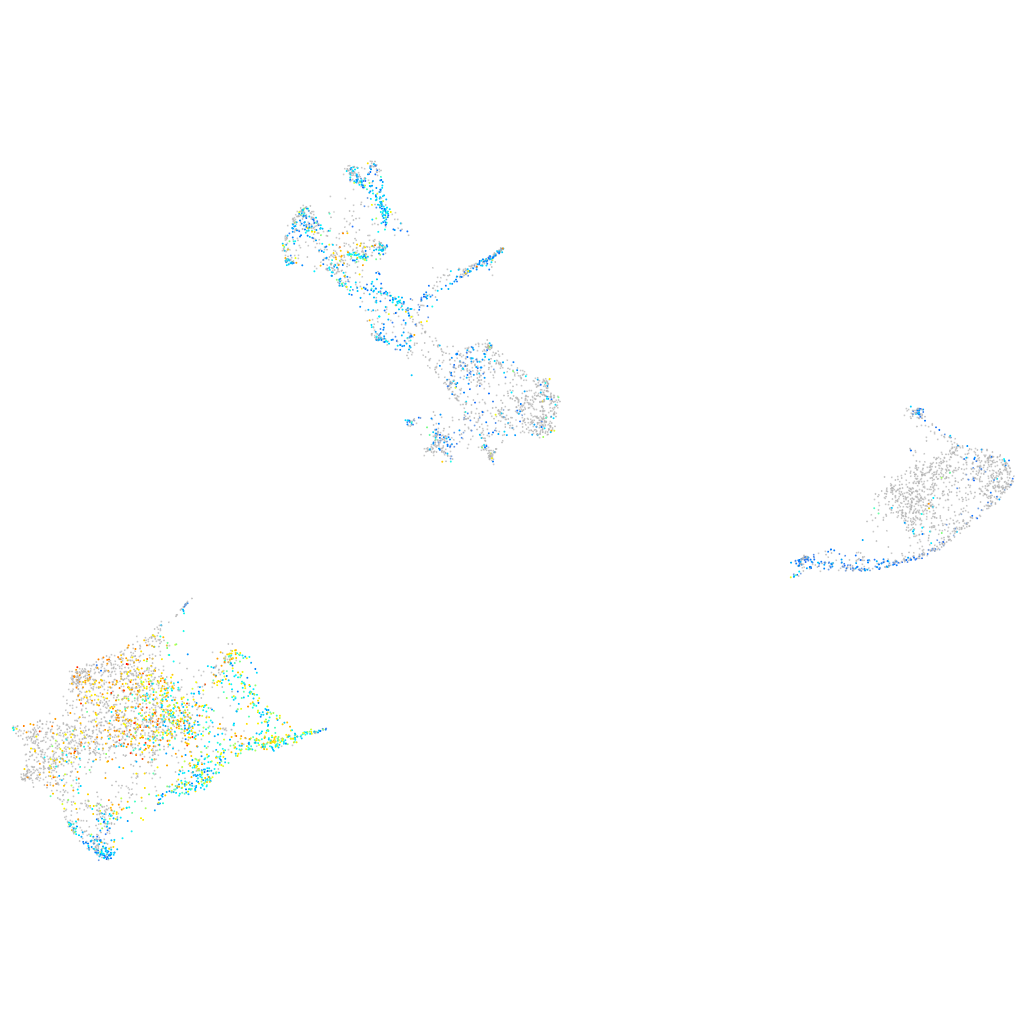
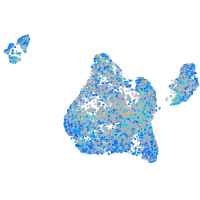
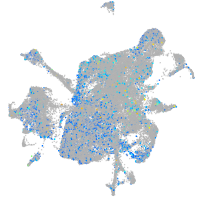
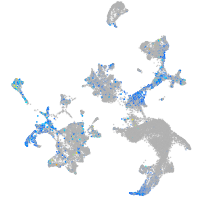
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| paics | 0.423 | tyrp1b | -0.286 |
| impdh1b | 0.413 | dct | -0.266 |
| glulb | 0.355 | zgc:91968 | -0.261 |
| prdx1 | 0.343 | tyrp1a | -0.261 |
| atic | 0.342 | pmela | -0.260 |
| sprb | 0.339 | icn | -0.245 |
| cx30.3 | 0.333 | oca2 | -0.241 |
| slc2a15a | 0.331 | prkar1b | -0.223 |
| cyb5a | 0.329 | s100a10b | -0.222 |
| akr1b1 | 0.318 | anxa1a | -0.215 |
| aox5 | 0.310 | si:ch73-389b16.1 | -0.210 |
| pts | 0.306 | mtbl | -0.204 |
| prps1a | 0.304 | slc24a5 | -0.202 |
| gmps | 0.304 | kita | -0.197 |
| uraha | 0.291 | slc22a2 | -0.171 |
| slc2a11b | 0.285 | slc39a10 | -0.167 |
| gart | 0.279 | aadac | -0.167 |
| CABZ01021592.1 | 0.275 | ckbb | -0.164 |
| tmem130 | 0.275 | spock3 | -0.158 |
| gpd1b | 0.268 | slc45a2 | -0.151 |
| prtfdc1 | 0.267 | slc29a3 | -0.149 |
| mdh1aa | 0.267 | ptmaa | -0.148 |
| cax1 | 0.262 | zdhhc2 | -0.144 |
| shmt1 | 0.261 | nova2 | -0.143 |
| sigmar1 | 0.259 | slc6a15 | -0.143 |
| pax7a | 0.257 | gpm6aa | -0.140 |
| mibp | 0.255 | CDK18 | -0.139 |
| rbp4l | 0.254 | slc30a1b | -0.139 |
| CABZ01032488.1 | 0.254 | yjefn3 | -0.139 |
| rgs2 | 0.251 | rap1gap | -0.137 |
| iah1 | 0.248 | hsd20b2 | -0.137 |
| mocs1 | 0.247 | stmn1b | -0.132 |
| si:dkey-251i10.2 | 0.246 | tyr | -0.132 |
| mocos | 0.243 | slc7a8a | -0.131 |
| sult1st1 | 0.243 | spra | -0.130 |