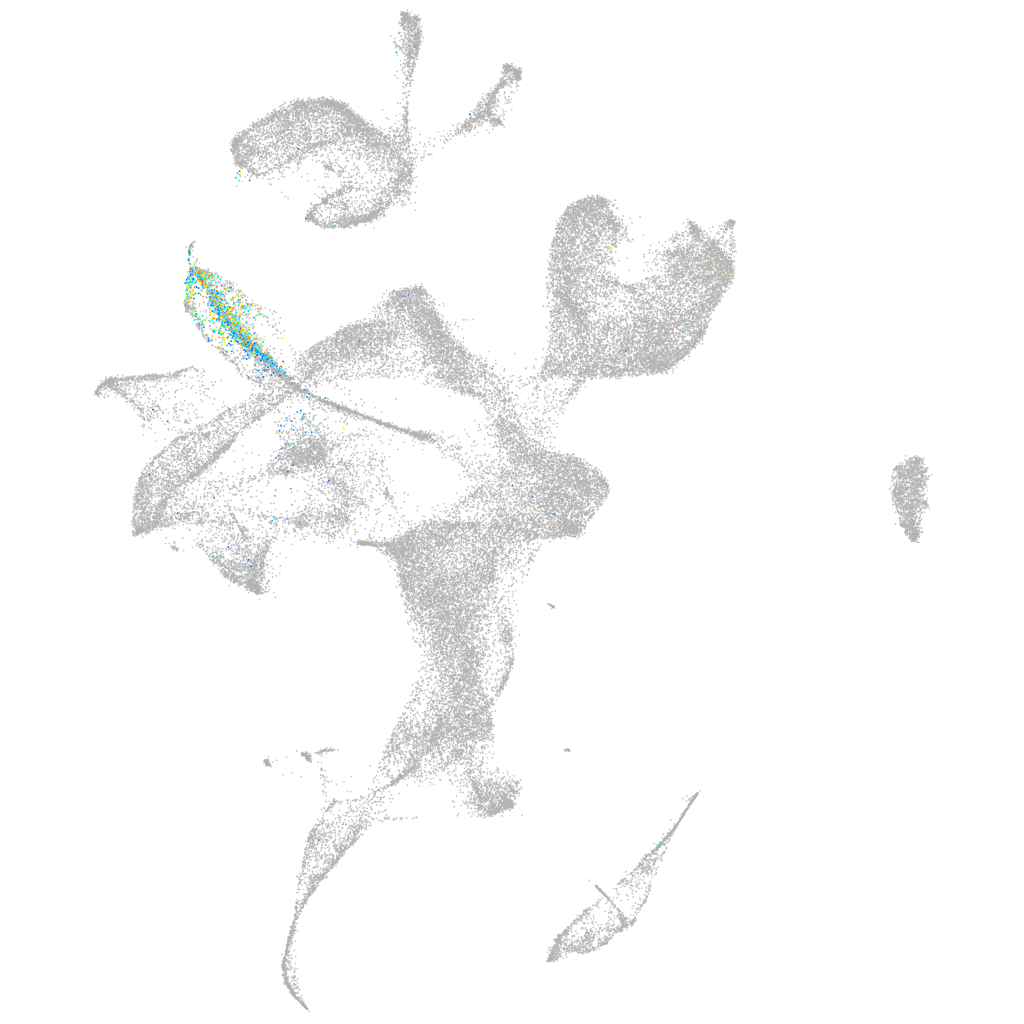
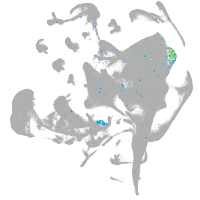
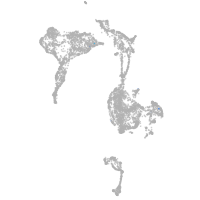
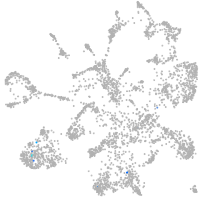
POU class 4 homeobox 3
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbpms2a | 0.548 | hmgb2a | -0.163 |
| pou4f2 | 0.544 | hmga1a | -0.094 |
| pou4f1 | 0.505 | mdka | -0.085 |
| rbpms2b | 0.431 | fabp7a | -0.085 |
| isl2b | 0.418 | msi1 | -0.083 |
| irx4a | 0.377 | ahcy | -0.081 |
| inab | 0.373 | pcna | -0.079 |
| oaz2b | 0.360 | stmn1a | -0.079 |
| islr2 | 0.348 | nutf2l | -0.075 |
| calb2a | 0.333 | dut | -0.074 |
| cplx2l | 0.332 | msna | -0.073 |
| cntn2 | 0.331 | rrm1 | -0.073 |
| BX247868.1 | 0.319 | mki67 | -0.072 |
| zgc:158291 | 0.315 | ccnd1 | -0.071 |
| nrn1a | 0.314 | lbr | -0.071 |
| stmn2a | 0.305 | selenoh | -0.070 |
| rab6ba | 0.304 | crx | -0.070 |
| grin2ab | 0.291 | neurod4 | -0.070 |
| irx2a | 0.290 | chaf1a | -0.070 |
| gap43 | 0.288 | anp32e | -0.069 |
| xpr1a | 0.283 | COX7A2 (1 of many) | -0.069 |
| eno2 | 0.281 | tuba8l | -0.069 |
| rtn1b | 0.279 | her15.1 | -0.068 |
| stmn4 | 0.276 | syt5b | -0.068 |
| stmn4l | 0.275 | si:ch73-281n10.2 | -0.068 |
| satb2 | 0.271 | otx5 | -0.067 |
| syt9b | 0.268 | ccna2 | -0.067 |
| nell2a | 0.267 | eef1da | -0.067 |
| syt2a | 0.266 | dek | -0.066 |
| ebf3a | 0.265 | anp32b | -0.065 |
| frem2a | 0.263 | cks1b | -0.065 |
| mllt11 | 0.259 | rpa3 | -0.064 |
| stmn2b | 0.258 | mcm7 | -0.064 |
| uchl1 | 0.256 | rpl5b | -0.064 |
| tkta | 0.255 | id1 | -0.063 |