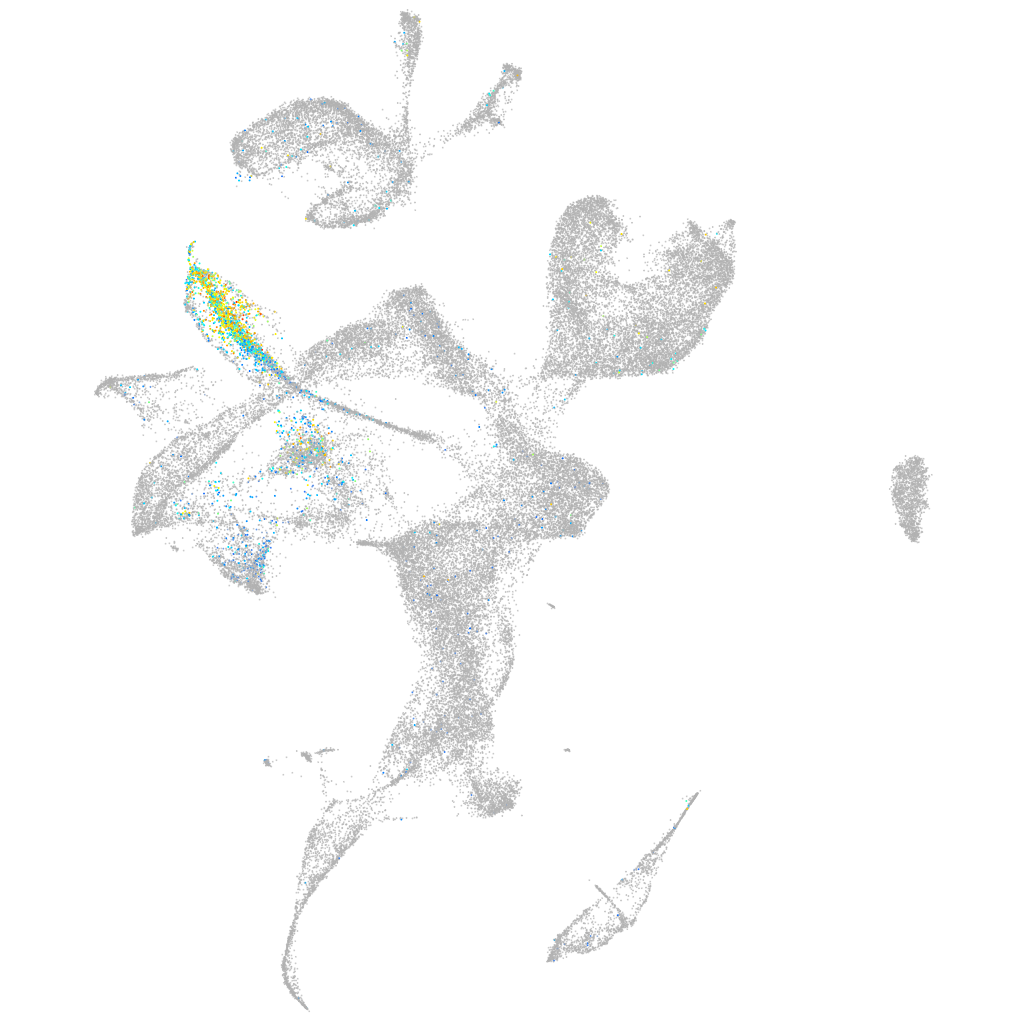
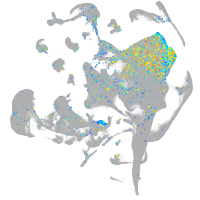
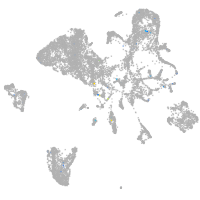
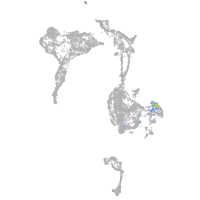
POU class 4 homeobox 1
ZFIN 
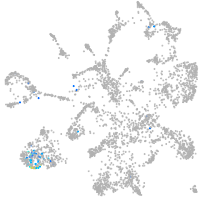
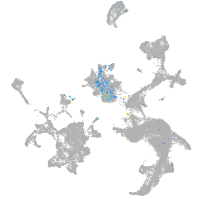
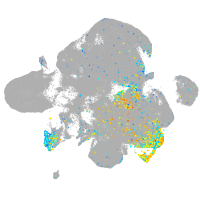
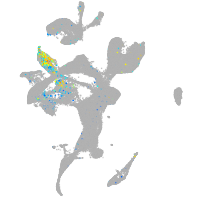
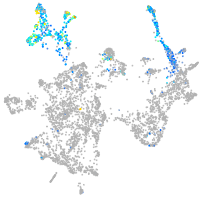
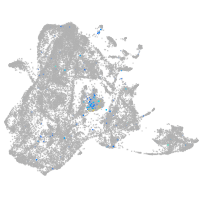
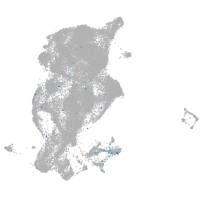
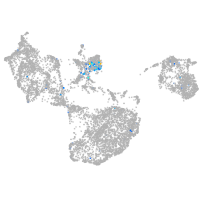
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbpms2a | 0.603 | hmgb2a | -0.199 |
| isl2b | 0.529 | pcna | -0.103 |
| pou4f2 | 0.518 | hmga1a | -0.103 |
| rbpms2b | 0.508 | ahcy | -0.102 |
| pou4f3 | 0.505 | mdka | -0.099 |
| zgc:158291 | 0.483 | msi1 | -0.098 |
| inab | 0.468 | anp32e | -0.097 |
| islr2 | 0.463 | nutf2l | -0.097 |
| nrn1a | 0.402 | stmn1a | -0.096 |
| BX247868.1 | 0.395 | rrm1 | -0.096 |
| cplx2l | 0.389 | fabp7a | -0.095 |
| cntn2 | 0.377 | dut | -0.094 |
| irx4a | 0.376 | ccnd1 | -0.092 |
| stmn2a | 0.370 | chaf1a | -0.091 |
| xpr1a | 0.368 | msna | -0.091 |
| frem2a | 0.364 | mki67 | -0.090 |
| oaz2b | 0.363 | selenoh | -0.090 |
| calb2a | 0.357 | lbr | -0.088 |
| grin2ab | 0.357 | COX7A2 (1 of many) | -0.086 |
| gap43 | 0.354 | tuba8l | -0.086 |
| stmn4 | 0.350 | ccna2 | -0.086 |
| rtn1b | 0.348 | lig1 | -0.085 |
| eno2 | 0.347 | si:ch73-281n10.2 | -0.084 |
| GNB4 | 0.346 | mcm7 | -0.084 |
| tmsb | 0.343 | rpa3 | -0.083 |
| ebf3a | 0.335 | dek | -0.083 |
| stmn4l | 0.334 | banf1 | -0.082 |
| syt2a | 0.332 | neurod4 | -0.082 |
| si:ch211-12e13.12 | 0.327 | cks1b | -0.081 |
| stmn2b | 0.327 | her15.1 | -0.081 |
| id4 | 0.321 | anp32b | -0.080 |
| rab6ba | 0.315 | CABZ01005379.1 | -0.080 |
| satb2 | 0.310 | rrm2 | -0.080 |
| kif26bb | 0.306 | mcm6 | -0.079 |
| mllt11 | 0.306 | mibp | -0.078 |