"polymerase (DNA directed), epsilon 2"
ZFIN 
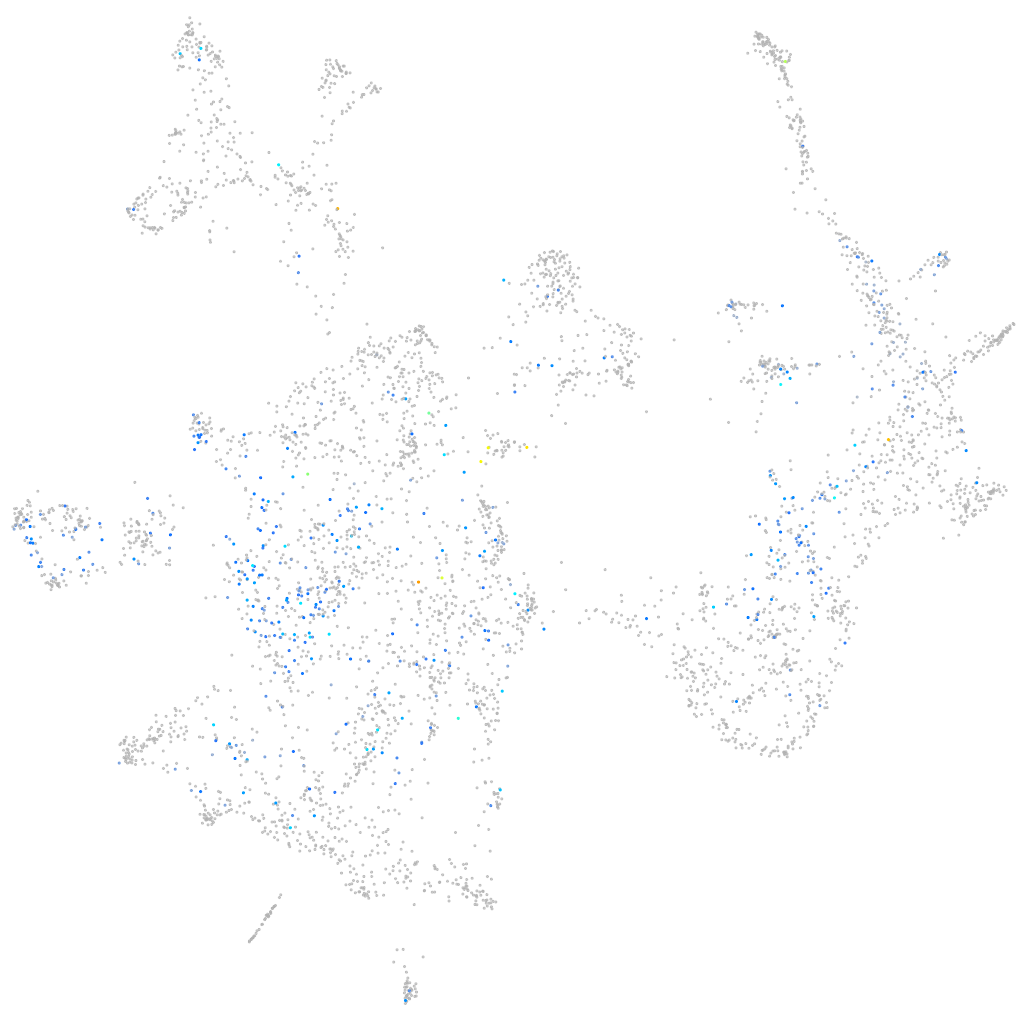
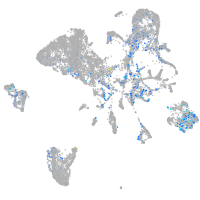
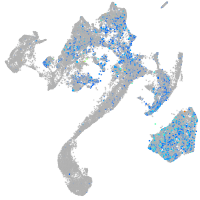
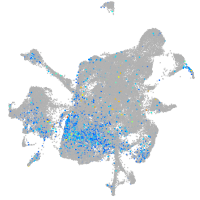
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.375 | h1f0 | -0.125 |
| dut | 0.362 | nupr1a | -0.112 |
| rrm2 | 0.357 | atp2b1a | -0.105 |
| rrm1 | 0.347 | tob1b | -0.098 |
| lig1 | 0.345 | tmem59 | -0.098 |
| zgc:110540 | 0.345 | gabarapb | -0.096 |
| chaf1a | 0.333 | ip6k2a | -0.096 |
| rpa3 | 0.333 | calm1a | -0.094 |
| stmn1a | 0.330 | ccni | -0.094 |
| prim2 | 0.325 | atp1a3b | -0.092 |
| rpa2 | 0.323 | rnasekb | -0.090 |
| CABZ01005379.1 | 0.323 | spint2 | -0.088 |
| fen1 | 0.322 | hist2h2l | -0.087 |
| nasp | 0.320 | calm1b | -0.085 |
| unga | 0.312 | tspan13b | -0.085 |
| mcm7 | 0.306 | cd164l2 | -0.085 |
| tk1 | 0.305 | calml4a | -0.085 |
| tyms | 0.304 | gabarapl2 | -0.085 |
| dek | 0.298 | nptna | -0.085 |
| slbp | 0.296 | emb | -0.085 |
| dnajc9 | 0.294 | btc | -0.084 |
| rfc4 | 0.294 | gdi1 | -0.083 |
| cks1b | 0.293 | mcl1a | -0.082 |
| esco2 | 0.293 | atp1b2b | -0.082 |
| pold1 | 0.288 | eno1a | -0.082 |
| si:dkey-6i22.5 | 0.287 | ckbb | -0.081 |
| kif15 | 0.283 | si:ch73-15n15.3 | -0.081 |
| mibp | 0.282 | gpx2 | -0.081 |
| dhfr | 0.281 | ube2h | -0.081 |
| chek1 | 0.278 | CR383676.1 | -0.081 |
| chaf1b | 0.276 | gapdhs | -0.080 |
| ube2t | 0.275 | tpi1a | -0.080 |
| asf1ba | 0.274 | ap3s1.1 | -0.079 |
| tubb2b | 0.274 | prr15la | -0.079 |
| msh2 | 0.272 | lrrc73 | -0.079 |