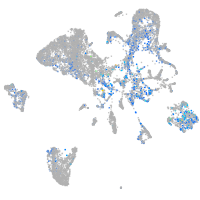
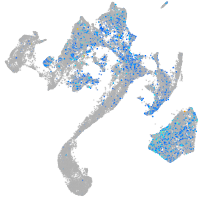
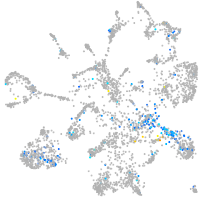
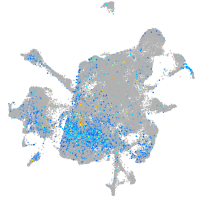
"polymerase (DNA directed), alpha 2"
ZFIN 
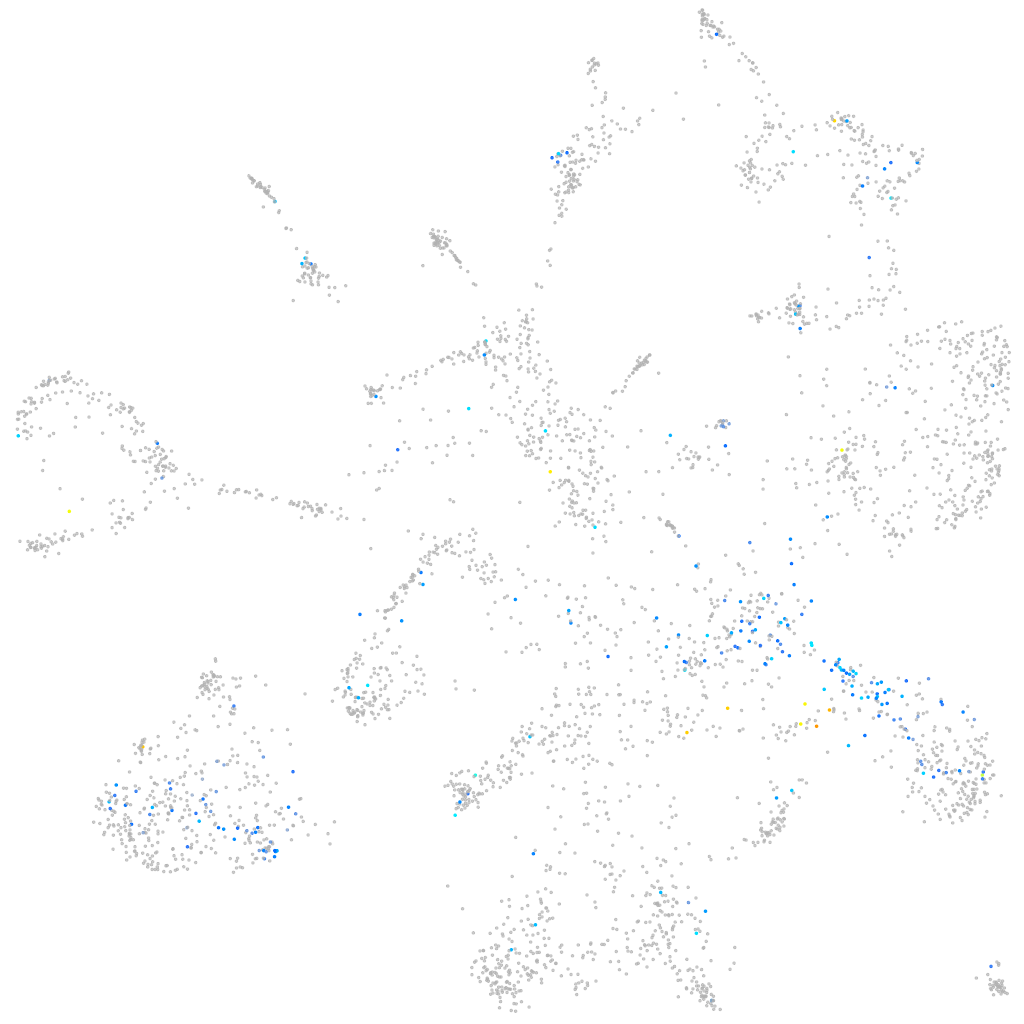
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.371 | cebpd | -0.089 |
| rrm1 | 0.351 | tpm1 | -0.087 |
| dut | 0.344 | ifitm1 | -0.078 |
| CABZ01005379.1 | 0.325 | tagln | -0.076 |
| chaf1a | 0.325 | lxn | -0.076 |
| slbp | 0.320 | zgc:162730 | -0.072 |
| rpa2 | 0.318 | mfap2 | -0.071 |
| zgc:110540 | 0.313 | dap1b | -0.071 |
| fen1 | 0.303 | acta2 | -0.071 |
| nasp | 0.289 | ppdpfb | -0.068 |
| rpa3 | 0.289 | CR383676.1 | -0.068 |
| rrm2 | 0.288 | tob1b | -0.068 |
| LOC100330864 | 0.282 | myl9a | -0.068 |
| tyms | 0.282 | ahnak | -0.067 |
| stmn1a | 0.279 | selenow1 | -0.066 |
| si:ch211-156b7.4 | 0.272 | krt8 | -0.065 |
| tk1 | 0.265 | tp53inp1 | -0.065 |
| mibp | 0.263 | cxcl14 | -0.064 |
| dhfr | 0.260 | zgc:92606 | -0.064 |
| chek1 | 0.260 | pvalb1 | -0.063 |
| sumo3b | 0.260 | actb2 | -0.063 |
| dck | 0.256 | pvalb2 | -0.063 |
| prim2 | 0.251 | nupr1a | -0.062 |
| lig1 | 0.250 | cavin2b | -0.061 |
| esco2 | 0.250 | lmod1b | -0.061 |
| asf1ba | 0.247 | zgc:153867 | -0.061 |
| XLOC-042899 | 0.243 | myl9b | -0.060 |
| chtf8 | 0.238 | tpm2 | -0.060 |
| dnajc9 | 0.233 | rbp4 | -0.059 |
| mcm7 | 0.230 | sparc | -0.059 |
| mis12 | 0.229 | pgm5 | -0.059 |
| atad2 | 0.229 | ctsla | -0.059 |
| cdc45 | 0.229 | tmsb4x | -0.059 |
| rbbp4 | 0.227 | tpm4b | -0.059 |
| gins2 | 0.226 | ckba | -0.058 |