"polymerase (DNA directed), alpha 2"
ZFIN 
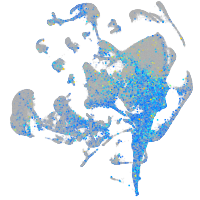
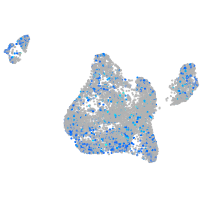
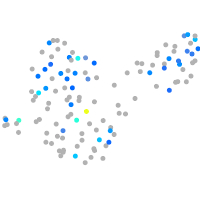
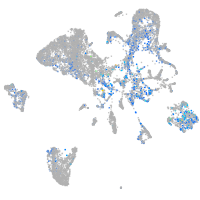
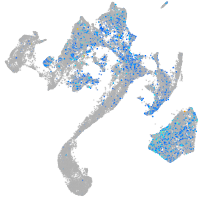
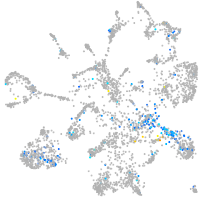
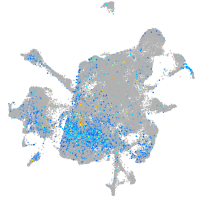
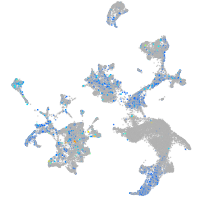
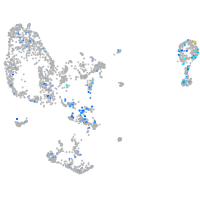
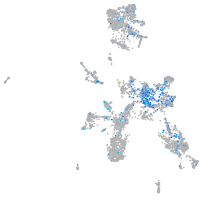
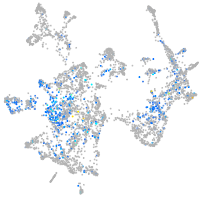
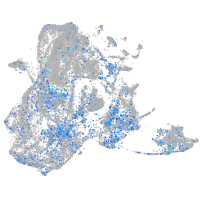
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.491 | BX000438.2 | -0.178 |
| pcna | 0.489 | selenow1 | -0.175 |
| slbp | 0.472 | ppdpfb | -0.163 |
| rrm2 | 0.452 | zgc:153284 | -0.162 |
| rpa2 | 0.451 | si:dkey-193p11.2 | -0.160 |
| dut | 0.446 | tob1b | -0.156 |
| mcm7 | 0.434 | si:ch211-195b11.3 | -0.154 |
| chaf1a | 0.432 | aldob | -0.151 |
| rrm1 | 0.426 | tdh | -0.147 |
| nasp | 0.426 | si:dkey-248g15.3 | -0.142 |
| rpa3 | 0.412 | abcb5 | -0.141 |
| fen1 | 0.410 | fbp1a | -0.139 |
| CABZ01005379.1 | 0.409 | soul2 | -0.138 |
| rfc3 | 0.404 | icn2 | -0.138 |
| LOC100330864 | 0.399 | si:dkey-247k7.2 | -0.136 |
| asf1ba | 0.385 | zgc:193505 | -0.135 |
| tk1 | 0.385 | icn | -0.134 |
| stmn1a | 0.385 | zgc:110333 | -0.134 |
| rbbp4 | 0.382 | mt-co2 | -0.133 |
| esco2 | 0.379 | nupr1a | -0.131 |
| dck | 0.378 | COX3 | -0.130 |
| dek | 0.373 | enosf1 | -0.130 |
| prim2 | 0.372 | ahnak | -0.127 |
| sumo3b | 0.369 | selenow2b | -0.127 |
| dhfr | 0.368 | mgst1.2 | -0.126 |
| lig1 | 0.360 | cst14b.1 | -0.125 |
| cdk2 | 0.359 | si:dkey-184p9.7 | -0.125 |
| si:ch211-156b7.4 | 0.357 | rnf183 | -0.124 |
| mcm6 | 0.355 | scel | -0.123 |
| mcm4 | 0.353 | epdl1 | -0.123 |
| mcm5 | 0.352 | gstp1 | -0.121 |
| gins2 | 0.352 | atp1b1b | -0.121 |
| e2f7 | 0.351 | ppl | -0.119 |
| znfl2a | 0.350 | sqstm1 | -0.118 |
| mibp | 0.350 | bloc1s6 | -0.118 |