pleiomorphic adenoma gene 1
ZFIN 
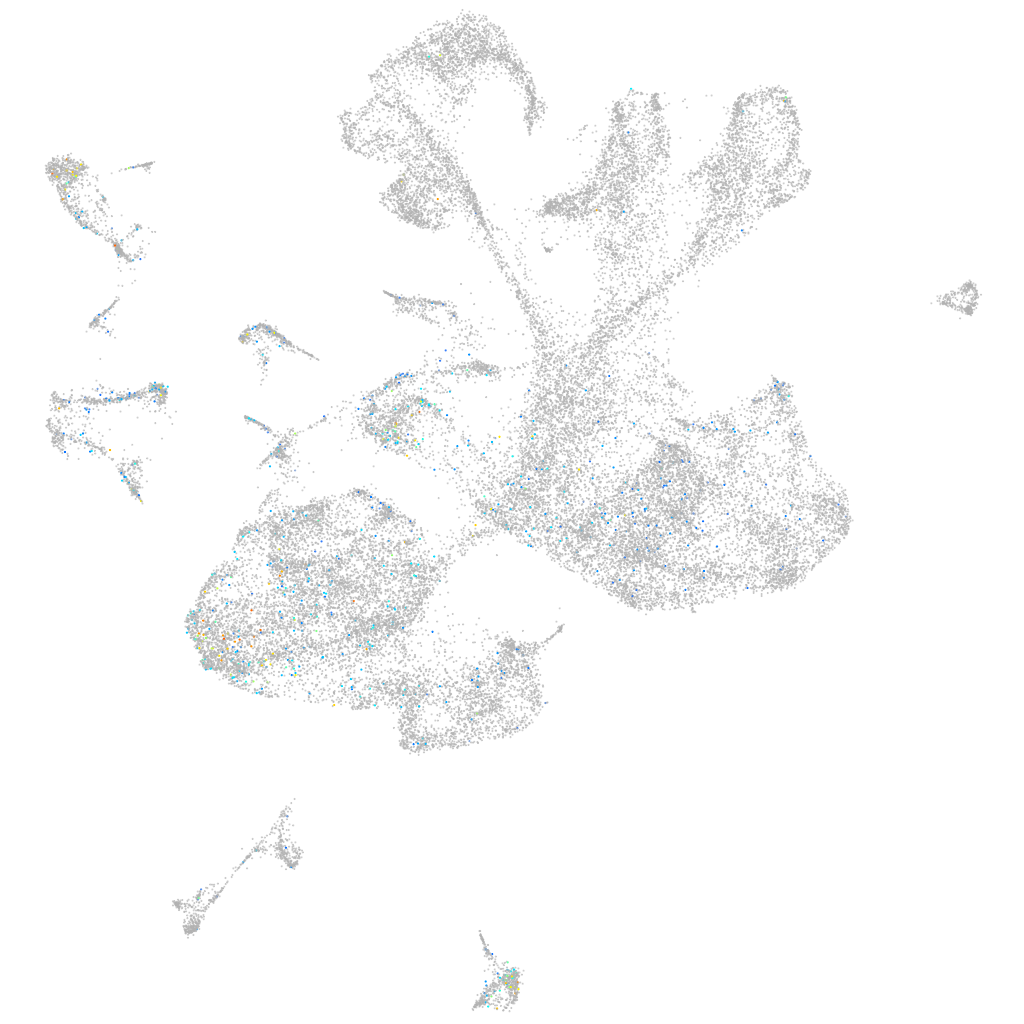
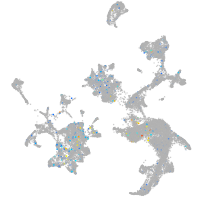
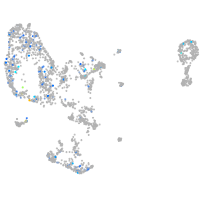
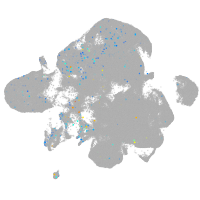
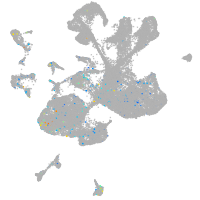
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43 | 0.109 | ptmab | -0.095 |
| ptn | 0.102 | si:ch211-222l21.1 | -0.084 |
| gpr37l1b | 0.102 | h3f3d | -0.083 |
| atp1a1b | 0.100 | tubb2b | -0.080 |
| slc3a2a | 0.098 | hnrnpaba | -0.078 |
| glula | 0.097 | hnrnpa0l | -0.076 |
| si:ch211-66e2.5 | 0.097 | khdrbs1a | -0.076 |
| ppap2d | 0.095 | elavl3 | -0.074 |
| slc4a4a | 0.095 | h3f3a | -0.072 |
| fabp7a | 0.095 | cirbpb | -0.071 |
| slc1a2b | 0.093 | h2afvb | -0.070 |
| cyp2ad3 | 0.091 | marcksb | -0.067 |
| efhd1 | 0.087 | hsp90ab1 | -0.066 |
| sparc | 0.084 | hmga1a | -0.066 |
| mt2 | 0.084 | hmgb1b | -0.064 |
| hepacama | 0.083 | hnrnpa0b | -0.064 |
| cebpd | 0.082 | rps27.1 | -0.064 |
| gpm6bb | 0.081 | rps23 | -0.064 |
| atp1b4 | 0.080 | si:ch73-1a9.3 | -0.062 |
| mlc1 | 0.079 | si:ch211-288g17.3 | -0.062 |
| eno1b | 0.079 | myt1b | -0.061 |
| cdo1 | 0.079 | cirbpa | -0.059 |
| cox4i2 | 0.078 | snrpd2 | -0.059 |
| cahz | 0.078 | hspa8 | -0.058 |
| qki2 | 0.078 | ran | -0.058 |
| gpc5b | 0.078 | hmgn2 | -0.058 |
| s1pr1 | 0.078 | tubb5 | -0.058 |
| or102-2 | 0.077 | sumo3a | -0.057 |
| FO704813.1 | 0.077 | hmgb2b | -0.057 |
| mboat1 | 0.076 | smarce1 | -0.057 |
| CR933734.1 | 0.076 | srsf3b | -0.057 |
| npc2 | 0.076 | chd4a | -0.056 |
| spry2 | 0.075 | snrpf | -0.056 |
| si:ch1073-303k11.2 | 0.075 | nfyba | -0.056 |
| acbd7 | 0.075 | zc4h2 | -0.055 |