praja ring finger ubiquitin ligase 2
ZFIN 
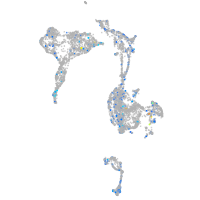
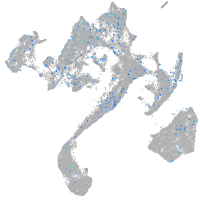
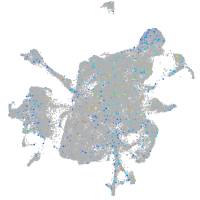
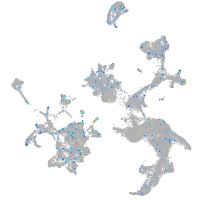
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR388079.2 | 0.148 | apoc1 | -0.061 |
| CR391910.2 | 0.138 | rbp2b | -0.058 |
| CU915256.2 | 0.137 | fabp10a | -0.057 |
| si:ch1073-268j14.1 | 0.136 | bhmt | -0.056 |
| RASGRF1 | 0.110 | ftcd | -0.055 |
| asb10 | 0.109 | agxtb | -0.055 |
| cacna1sb | 0.108 | apoa1b | -0.054 |
| XLOC-014193 | 0.107 | grhprb | -0.053 |
| stx3a | 0.106 | apoa2 | -0.052 |
| KLHL28 | 0.105 | apom | -0.052 |
| capns1a | 0.104 | mat1a | -0.052 |
| slc35f2l | 0.102 | rbp4 | -0.051 |
| si:cabz01007812.1 | 0.097 | ttr | -0.051 |
| pkma | 0.097 | aqp12 | -0.051 |
| capns1b | 0.097 | uox | -0.050 |
| zgc:194007 | 0.096 | hpda | -0.050 |
| oclnb | 0.095 | LOC110437731 | -0.049 |
| gdpd3a | 0.094 | ambp | -0.049 |
| phlda2 | 0.093 | fgb | -0.048 |
| agr2 | 0.092 | comtd1 | -0.048 |
| cd63 | 0.092 | zgc:123103 | -0.048 |
| capn2b | 0.092 | gamt | -0.048 |
| icn | 0.091 | si:dkey-16p21.8 | -0.047 |
| s100v1 | 0.090 | gc | -0.047 |
| trim69 | 0.090 | agxta | -0.046 |
| tmsb1 | 0.090 | serpina1l | -0.046 |
| dnah6 | 0.089 | ttc36 | -0.046 |
| gnb1a | 0.089 | f7i | -0.046 |
| ier2b | 0.089 | gapdh | -0.046 |
| tprg1 | 0.089 | hao1 | -0.046 |
| zgc:158343 | 0.089 | c9 | -0.046 |
| micu2 | 0.088 | hgd | -0.045 |
| si:dkey-106l3.7 | 0.088 | fetub | -0.045 |
| atf4b | 0.088 | serpina1 | -0.045 |
| CU019562.3 | 0.088 | si:dkey-86l18.10 | -0.044 |