piwi-like RNA-mediated gene silencing 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory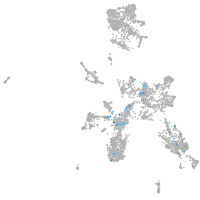 |
otic / lateral line |
epidermis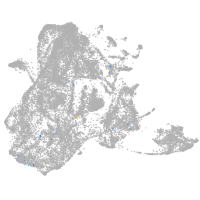 |
periderm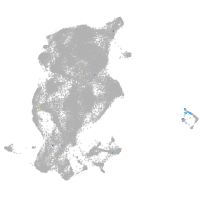 |
fin |
ionocytes / mucous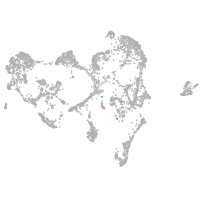 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:171500 | 0.163 | CR383676.1 | -0.030 |
| si:dkey-203a12.5 | 0.137 | tmsb | -0.028 |
| LO018315.7 | 0.115 | gpm6aa | -0.025 |
| CR352226.2 | 0.112 | fabp3 | -0.025 |
| il19l | 0.110 | rtn1a | -0.025 |
| CR788249.1 | 0.087 | elavl3 | -0.024 |
| LOC101886779 | 0.081 | actc1b | -0.023 |
| gig2i | 0.078 | marcksl1b | -0.023 |
| drd7 | 0.075 | tuba1c | -0.023 |
| BX927258.1 | 0.075 | myt1b | -0.023 |
| XLOC-022683 | 0.074 | tmeff1b | -0.022 |
| hspb1 | 0.071 | pvalb1 | -0.022 |
| ftr92 | 0.070 | pvalb2 | -0.022 |
| BX664750.2 | 0.068 | gnao1a | -0.021 |
| apela | 0.067 | elavl4 | -0.021 |
| BX001047.1 | 0.065 | gng3 | -0.021 |
| XLOC-032526 | 0.064 | sncb | -0.020 |
| chrd | 0.062 | marcksl1a | -0.020 |
| fgf8a | 0.062 | nova2 | -0.020 |
| LOC101886083 | 0.061 | tmsb4x | -0.020 |
| nfkb1 | 0.061 | si:dkey-276j7.1 | -0.020 |
| sp5l | 0.059 | ywhag2 | -0.020 |
| si:dkey-266f7.5 | 0.059 | gatad2b | -0.019 |
| cdx4 | 0.059 | gng2 | -0.019 |
| CR954963.1 | 0.056 | vim | -0.019 |
| si:dkey-48g21.7 | 0.055 | ube2d4 | -0.019 |
| LOC101884251 | 0.054 | ccni | -0.019 |
| nop58 | 0.052 | stx1b | -0.018 |
| XLOC-039121 | 0.052 | id4 | -0.018 |
| CU463284.2 | 0.050 | alcama | -0.018 |
| npm1a | 0.049 | ppdpfb | -0.018 |
| lrrc17 | 0.049 | gabarapl2 | -0.018 |
| si:ch73-315f9.2 | 0.049 | ckbb | -0.018 |
| BX005254.3 | 0.048 | si:dkeyp-75h12.5 | -0.018 |
| XLOC-001603 | 0.048 | appa | -0.018 |




