phosphatidylinositol transfer protein cytoplasmic 1b
ZFIN 
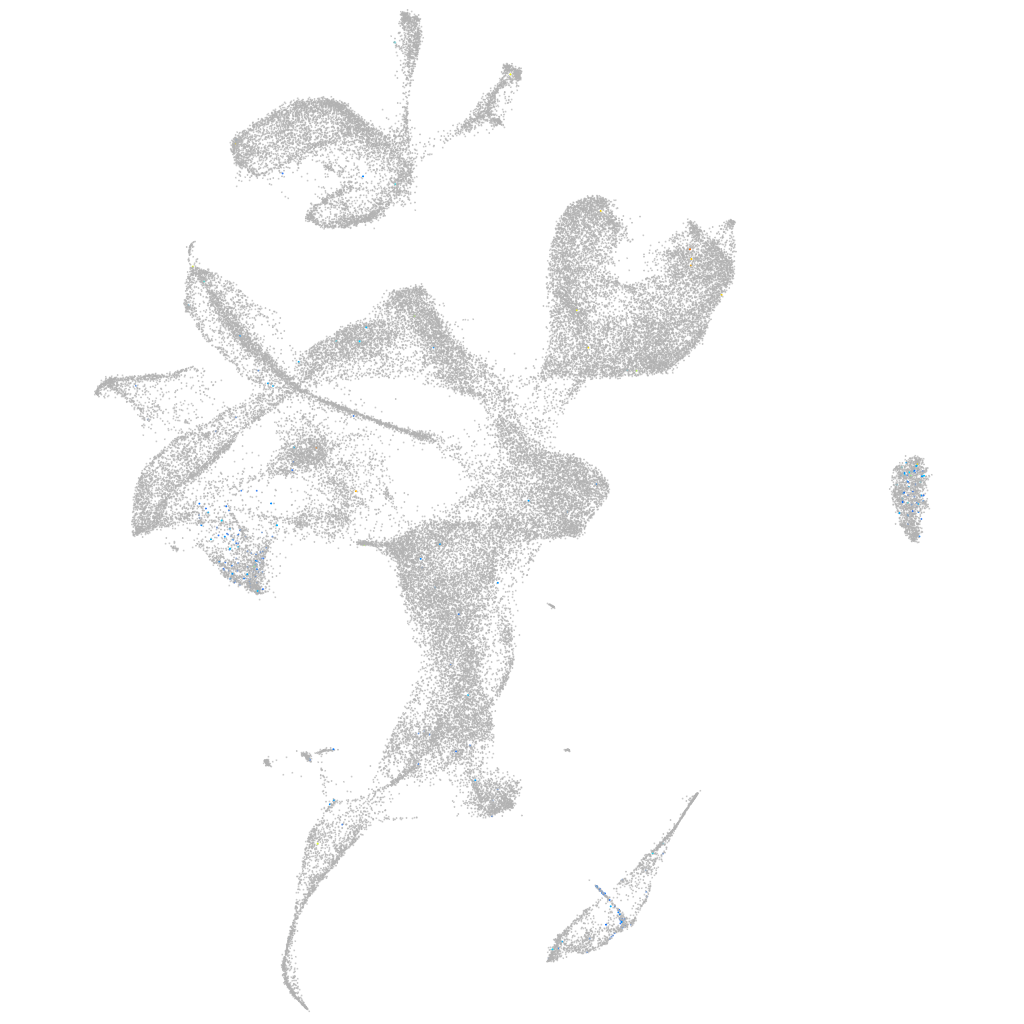
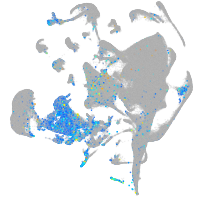
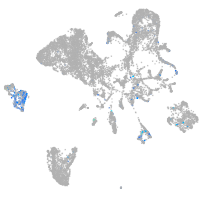
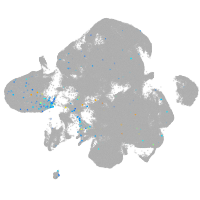
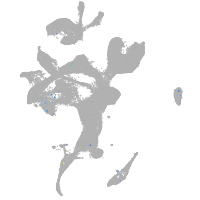
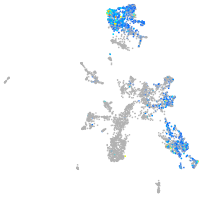
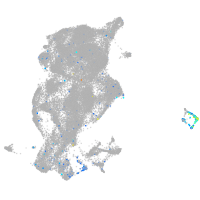
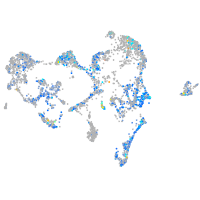
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| trim35-19 | 0.135 | h3f3a | -0.032 |
| FQ377991.1 | 0.111 | cadm3 | -0.031 |
| FP103048.1 | 0.082 | rps10 | -0.028 |
| CU929094.2 | 0.081 | ppiab | -0.026 |
| XLOC-033087 | 0.079 | hnrnpa0l | -0.026 |
| CABZ01118270.1 | 0.075 | fabp7a | -0.025 |
| fem1a | 0.067 | tuba1a | -0.025 |
| CU694486.1 | 0.064 | ptmaa | -0.024 |
| si:ch211-152c2.3 | 0.062 | rorb | -0.023 |
| col1a2 | 0.061 | COX3 | -0.023 |
| CR352226.6 | 0.061 | mt-atp6 | -0.023 |
| epcam | 0.061 | fabp3 | -0.023 |
| dnase1l4.1 | 0.059 | sinhcafl | -0.022 |
| nr6a1a | 0.059 | tuba1c | -0.022 |
| muc5f | 0.058 | rpl37 | -0.022 |
| hspb1 | 0.058 | gpm6aa | -0.021 |
| atp1b1a | 0.058 | ptmab | -0.020 |
| CU929178.1 | 0.057 | zgc:158463 | -0.020 |
| nitr1m | 0.057 | foxg1b | -0.019 |
| krt8 | 0.057 | mt-co2 | -0.019 |
| rerglb | 0.056 | zgc:114188 | -0.019 |
| apoeb | 0.055 | neurod4 | -0.019 |
| lama2 | 0.055 | hes6 | -0.018 |
| zgc:162730 | 0.055 | gpm6ab | -0.018 |
| si:ch73-335l21.4 | 0.054 | zgc:153409 | -0.017 |
| dlx3b | 0.054 | hmgn2 | -0.017 |
| cd9b | 0.054 | foxn4 | -0.017 |
| krt18a.1 | 0.054 | otx5 | -0.017 |
| pmp22a | 0.053 | insm1a | -0.016 |
| LOC103911701 | 0.053 | vsx2 | -0.016 |
| f11r.1 | 0.052 | bcl2l10 | -0.016 |
| stm | 0.052 | rps17 | -0.016 |
| sox1a | 0.052 | golga7ba | -0.016 |
| XLOC-042928 | 0.051 | epb41a | -0.016 |
| col6a4a | 0.051 | acbd7 | -0.016 |