phosphatidylinositol 4-kinase type 2 alpha
ZFIN 
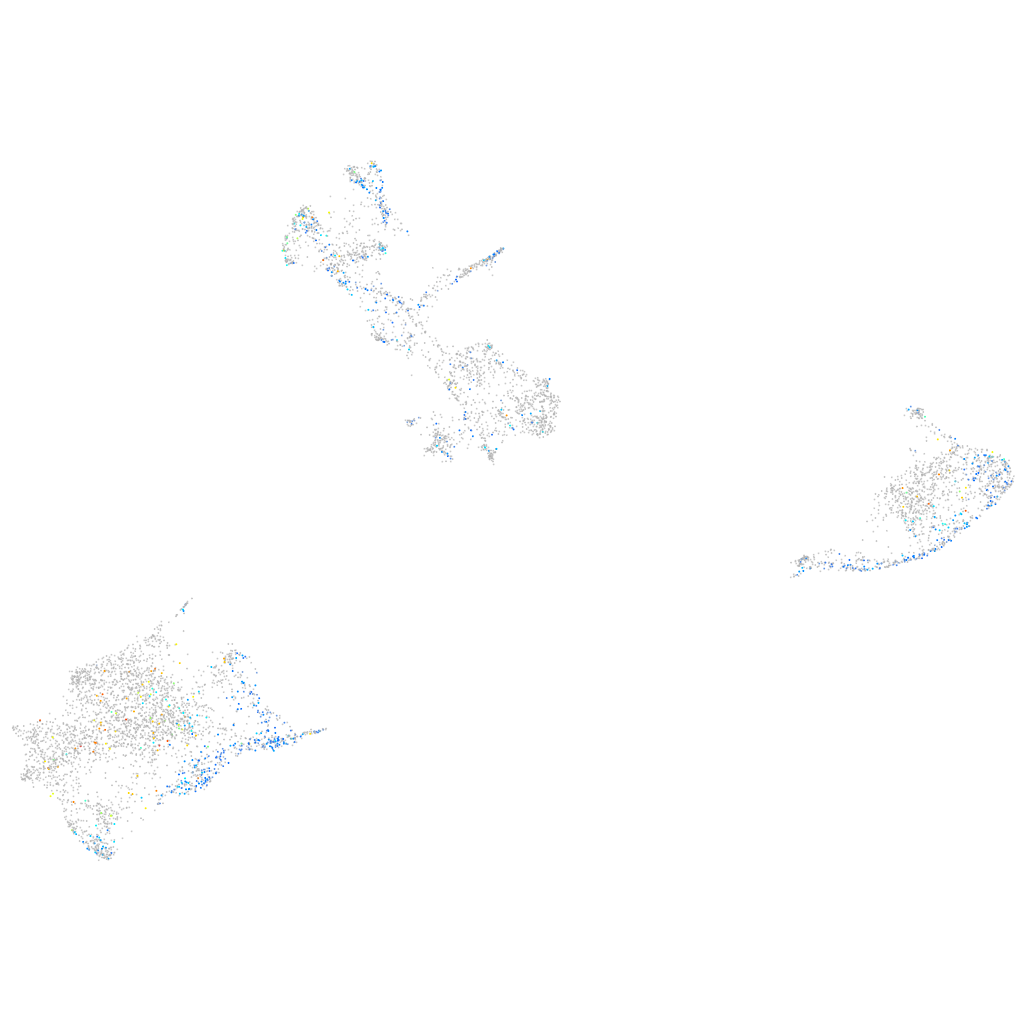
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gmps | 0.106 | ptmaa | -0.078 |
| si:zfos-943e10.1 | 0.105 | hbbe1.3 | -0.072 |
| slc2a15a | 0.105 | stmn1b | -0.069 |
| BX908804.2 | 0.104 | hbae1.1 | -0.067 |
| rnaseka | 0.103 | si:ch211-222l21.1 | -0.064 |
| rgs3a | 0.103 | elavl3 | -0.064 |
| pax7a | 0.100 | hbae3 | -0.064 |
| sox10 | 0.100 | pvalb1 | -0.060 |
| zgc:110239 | 0.099 | pvalb2 | -0.059 |
| trpm1b | 0.098 | epb41a | -0.058 |
| atp11a | 0.097 | hmgb1b | -0.058 |
| LOC103910009 | 0.096 | gpm6aa | -0.057 |
| atic | 0.095 | actc1b | -0.057 |
| glulb | 0.095 | nova2 | -0.056 |
| id3 | 0.093 | tuba1c | -0.056 |
| XLOC-019062 | 0.093 | myhz1.1 | -0.056 |
| ddit3 | 0.093 | hmgb3a | -0.054 |
| sinup | 0.091 | tmsb | -0.054 |
| syngr1a | 0.091 | mylpfa | -0.052 |
| cfap43 | 0.090 | marcksl1a | -0.049 |
| glud1b | 0.089 | tmeff1b | -0.049 |
| ccdc22 | 0.089 | hmgn2 | -0.048 |
| pax7b | 0.088 | fabp7a | -0.048 |
| cx30.3 | 0.088 | gng3 | -0.048 |
| XLOC-012561 | 0.087 | her15.1 | -0.048 |
| CU915256.2 | 0.087 | sox11b | -0.048 |
| gart | 0.086 | zc4h2 | -0.047 |
| rab32a | 0.086 | mab21l2 | -0.047 |
| tmc4 | 0.086 | si:ch73-1a9.3 | -0.047 |
| inka1a | 0.086 | ptmab | -0.047 |
| opn5 | 0.086 | myhz1.2 | -0.046 |
| xbp1 | 0.086 | nat8l | -0.046 |
| XLOC-003678 | 0.085 | gng2 | -0.046 |
| cdk15 | 0.085 | hbbe1.1 | -0.045 |
| dera | 0.085 | scrt2 | -0.045 |