PHD finger protein 21Aa
ZFIN 
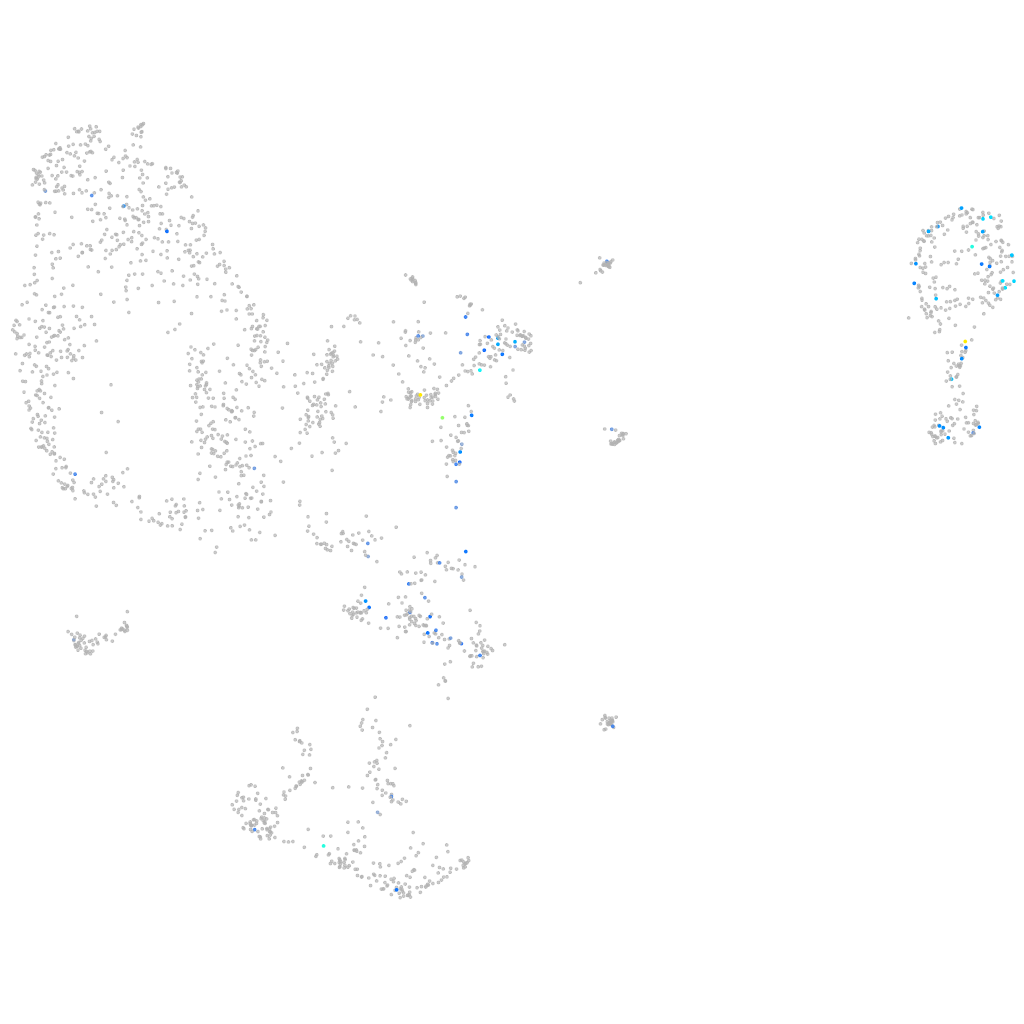
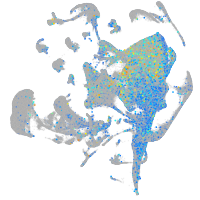
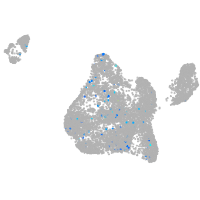
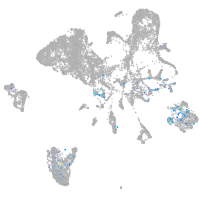
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-005215 | 0.258 | atp5f1b | -0.174 |
| LOC110439623 | 0.250 | atp1b1a | -0.164 |
| XLOC-034734 | 0.245 | eno3 | -0.158 |
| nefma | 0.238 | atp5pf | -0.151 |
| zgc:171482 | 0.232 | cox8a | -0.149 |
| LOC108190395 | 0.224 | rpl37 | -0.148 |
| si:ch1073-340i21.2 | 0.223 | suclg1 | -0.147 |
| pkn1b | 0.219 | atp5l | -0.146 |
| si:dkey-31f5.11 | 0.213 | cox5aa | -0.146 |
| CR848757.1 | 0.213 | atp5meb | -0.146 |
| dram1 | 0.211 | atp5mc3b | -0.144 |
| marcksb | 0.211 | atp1a1a.4 | -0.144 |
| bsx | 0.194 | elovl1b | -0.142 |
| LOC101883563 | 0.194 | ndrg1a | -0.140 |
| sncaip | 0.193 | si:ch211-235e9.6 | -0.139 |
| mipa | 0.190 | COX5B | -0.139 |
| chd7 | 0.188 | cox7c | -0.138 |
| syncrip | 0.187 | cdh17 | -0.138 |
| top1l | 0.186 | eif4a1b | -0.137 |
| uckl1a | 0.186 | slc25a5 | -0.136 |
| ccna2 | 0.186 | atp5mf | -0.135 |
| gipc3 | 0.186 | COX3 | -0.135 |
| seta | 0.185 | atp5mc1 | -0.134 |
| si:ch211-137a8.4 | 0.185 | rps10 | -0.134 |
| XPO5 | 0.183 | ndufa6 | -0.133 |
| hnrnpa1a | 0.183 | atp5f1d | -0.132 |
| marcksl1b | 0.182 | atp5f1e | -0.130 |
| nucks1a | 0.179 | mt-nd1 | -0.130 |
| akap12b | 0.179 | zgc:56493 | -0.130 |
| nkain1 | 0.178 | tpi1b | -0.129 |
| BX649620.1 | 0.178 | ppifb | -0.129 |
| asz1 | 0.177 | aldh6a1 | -0.127 |
| cyp46a1.3 | 0.176 | mdh1aa | -0.127 |
| si:ch73-1a9.3 | 0.175 | si:ch211-139a5.9 | -0.127 |
| zgc:113886 | 0.174 | ezra | -0.126 |