protein kinase N1b
ZFIN 
Other cell groups


all cells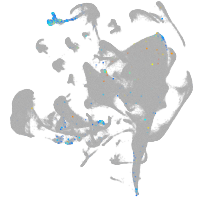 |
blastomeres |
PGCs |
endoderm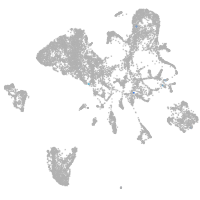 |
axial mesoderm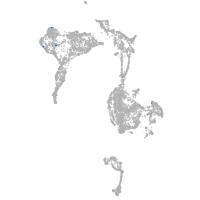 |
muscle  |
mural cells / non-skeletal muscle 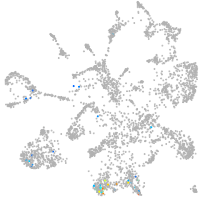 |
mesenchyme 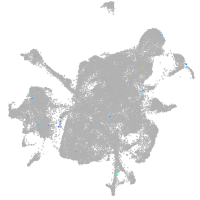 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia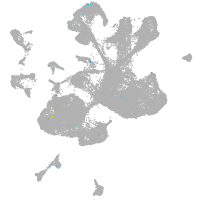 |
eye |
taste / olfactory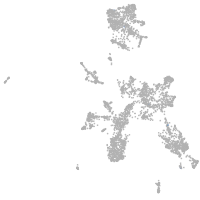 |
otic / lateral line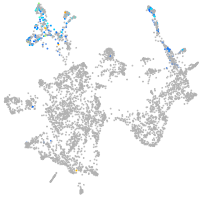 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| laptm5 | 0.330 | hmgb1b | -0.039 |
| coro1a | 0.304 | mdka | -0.039 |
| spi1b | 0.295 | nova2 | -0.035 |
| itgae.2 | 0.294 | si:ch211-137a8.4 | -0.033 |
| wasb | 0.285 | hmgb3a | -0.032 |
| FERMT3 (1 of many) | 0.285 | marcksb | -0.030 |
| zgc:64051 | 0.285 | tspan7 | -0.030 |
| ptprc | 0.283 | pfn2l | -0.029 |
| ccr9a | 0.281 | gpm6aa | -0.028 |
| fcer1gl | 0.277 | si:ch73-46j18.5 | -0.028 |
| CU499330.1 | 0.274 | her15.1 | -0.028 |
| glipr1a | 0.274 | id1 | -0.027 |
| grap2b | 0.274 | si:ch211-288g17.3 | -0.027 |
| itgb2 | 0.274 | sparc | -0.027 |
| lcp1 | 0.273 | cx43.4 | -0.026 |
| myo1f | 0.272 | hnrnpa0a | -0.026 |
| cxcr3.2 | 0.270 | pbx4 | -0.026 |
| si:ch211-102c2.4 | 0.266 | si:ch211-133n4.4 | -0.026 |
| arhgap15 | 0.262 | tmeff1b | -0.026 |
| plek | 0.262 | tuba1c | -0.026 |
| rgs13 | 0.262 | foxp4 | -0.025 |
| si:ch73-248e21.5 | 0.262 | mdkb | -0.025 |
| wasa | 0.262 | fabp3 | -0.024 |
| tnfaip8l2a | 0.262 | XLOC-003690 | -0.024 |
| LOC100536213 | 0.261 | cldn5a | -0.023 |
| spi1a | 0.260 | fabp7a | -0.023 |
| gmfg | 0.258 | FO082781.1 | -0.023 |
| si:ch211-243a20.4 | 0.258 | msi1 | -0.023 |
| gpr183a | 0.257 | apex1 | -0.022 |
| nrros | 0.256 | boc | -0.022 |
| lpar5a | 0.256 | efnb3b | -0.022 |
| adgrg3 | 0.254 | fam168a | -0.022 |
| parvg | 0.254 | rtn1a | -0.022 |
| csf1rb | 0.251 | sox11a | -0.022 |
| hmha1b | 0.250 | XLOC-003689 | -0.022 |




