PHD finger protein 10
ZFIN 
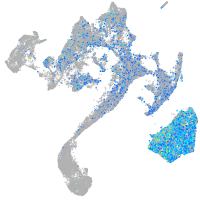
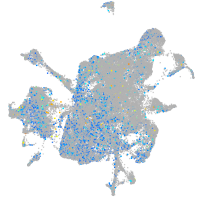
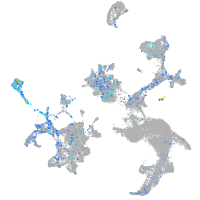
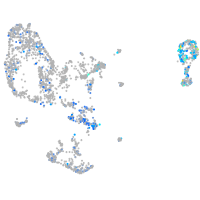
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152c2.3 | 0.207 | rpl37 | -0.142 |
| hspb1 | 0.194 | rps10 | -0.128 |
| stm | 0.194 | zgc:114188 | -0.116 |
| apoeb | 0.189 | h3f3a | -0.105 |
| NC-002333.4 | 0.181 | CR383676.1 | -0.104 |
| hnrnpub | 0.179 | ptmaa | -0.100 |
| hnrnpa1b | 0.178 | cspg5a | -0.100 |
| crabp2b | 0.177 | gapdhs | -0.098 |
| pou5f3 | 0.176 | zgc:158463 | -0.097 |
| rbm4.3 | 0.176 | pvalb2 | -0.092 |
| polr3gla | 0.170 | actc1b | -0.091 |
| apoc1 | 0.164 | atp6v0cb | -0.090 |
| si:dkey-66i24.9 | 0.164 | pvalb1 | -0.090 |
| s100a1 | 0.164 | calm1b | -0.089 |
| hnrnpa1a | 0.163 | marcksl1a | -0.085 |
| ilf3b | 0.163 | ppiab | -0.084 |
| zmp:0000000624 | 0.159 | mt-nd1 | -0.083 |
| seta | 0.158 | sv2a | -0.082 |
| hmga1a | 0.158 | tpi1b | -0.082 |
| nucks1a | 0.157 | nptna | -0.081 |
| nr6a1a | 0.156 | atp6ap2 | -0.078 |
| syncrip | 0.154 | zgc:56493 | -0.077 |
| fbl | 0.154 | mt-atp6 | -0.077 |
| khdrbs1a | 0.154 | sypb | -0.077 |
| anp32e | 0.154 | slc6a1a | -0.077 |
| hnrnpm | 0.153 | scg2b | -0.076 |
| COX7A2 | 0.151 | sncgb | -0.076 |
| bms1 | 0.151 | rnasekb | -0.076 |
| snrnp70 | 0.151 | snap25a | -0.076 |
| apela | 0.151 | aldocb | -0.074 |
| rbmx2 | 0.150 | tmsb4x | -0.074 |
| top1l | 0.150 | ywhag2 | -0.074 |
| ppig | 0.150 | calm1a | -0.074 |
| hnrnpabb | 0.149 | rps17 | -0.074 |
| ncl | 0.149 | eno1a | -0.073 |