phosphoglycerate kinase 1
ZFIN 
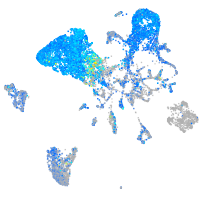
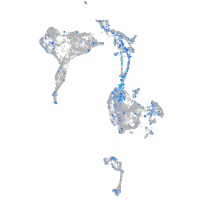
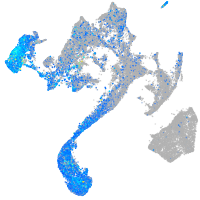
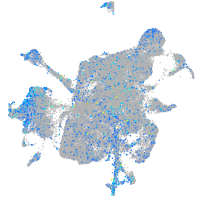
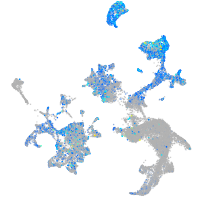
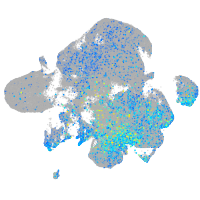
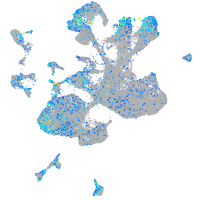
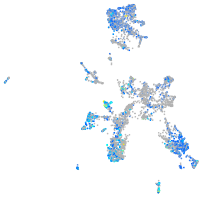
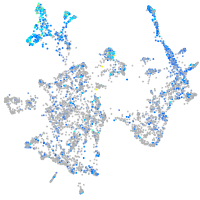
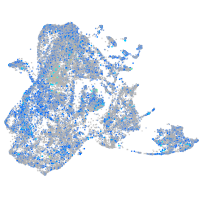
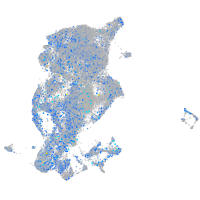
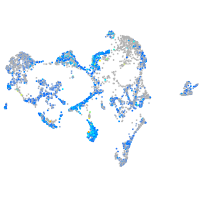
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1a | 0.500 | marcksb | -0.461 |
| gpia | 0.475 | anp32e | -0.450 |
| gapdhs | 0.458 | NC-002333.4 | -0.434 |
| serpinb1 | 0.446 | ncl | -0.413 |
| taf12 | 0.442 | syncrip | -0.412 |
| ppp1cbl | 0.432 | elavl1a | -0.410 |
| rnasekb | 0.431 | apoeb | -0.407 |
| ssu72 | 0.425 | si:ch73-281n10.2 | -0.407 |
| cd63 | 0.423 | ppig | -0.405 |
| zgc:56493 | 0.415 | zmat2 | -0.386 |
| cdkn1bb | 0.404 | hnrnpa1b | -0.381 |
| rpl14 | 0.402 | pnn | -0.364 |
| cox6a1 | 0.401 | fth1a | -0.361 |
| ccni | 0.395 | snrpb2 | -0.345 |
| rfx5 | 0.395 | ranbp1 | -0.345 |
| pigbos1 | 0.392 | ccna2 | -0.340 |
| hint2 | 0.391 | h2afva | -0.334 |
| mfap3l | 0.390 | hnrnpabb | -0.333 |
| cdr2l | 0.390 | khdrbs1a | -0.332 |
| tmem59 | 0.388 | hnrnpm | -0.331 |
| zgc:195170 | 0.387 | nono | -0.329 |
| prkab1b | 0.387 | hmga1a | -0.326 |
| tpma | 0.387 | ilf3b | -0.325 |
| st7 | 0.387 | dlgap5 | -0.322 |
| sccpdhb | 0.385 | fbl | -0.319 |
| vldlr | 0.385 | stm | -0.316 |
| rpl37 | 0.385 | hnrnpa1a | -0.315 |
| si:dkeyp-75h12.5 | 0.381 | zgc:77056 | -0.311 |
| cacna1da | 0.379 | akap12b | -0.311 |
| RAMP1 | 0.379 | tpx2 | -0.311 |
| kalrna | 0.378 | hnrnpub | -0.310 |
| tpi1b | 0.378 | si:ch211-288g17.3 | -0.309 |
| gsta.1 | 0.378 | ddx5 | -0.305 |
| etfa | 0.376 | npm1a | -0.305 |
| NC-002333.17 | 0.374 | nop58 | -0.304 |