phosphoglycerate kinase 1
ZFIN 
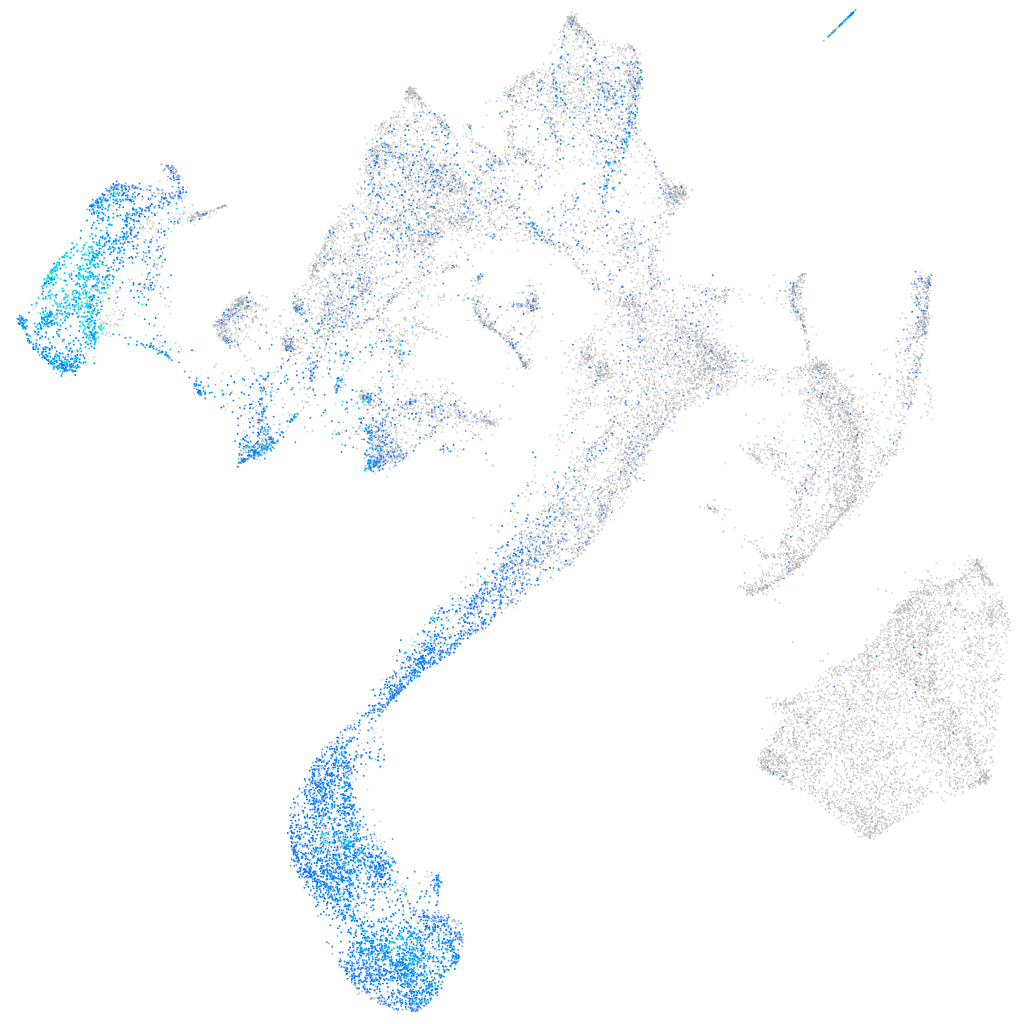
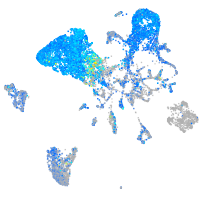
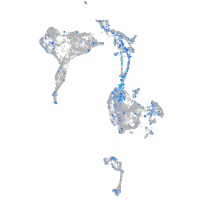
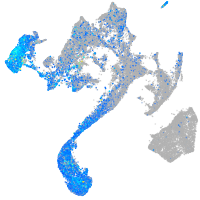
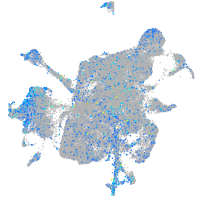
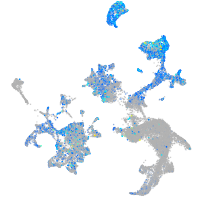
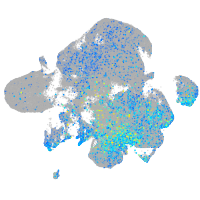
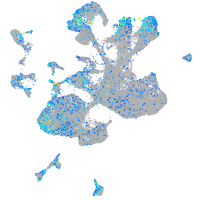
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdh | 0.711 | hsp90ab1 | -0.638 |
| pgam2 | 0.707 | h3f3d | -0.615 |
| eno3 | 0.702 | pabpc1a | -0.615 |
| ckmb | 0.697 | h2afvb | -0.591 |
| ckma | 0.696 | si:ch73-1a9.3 | -0.585 |
| aldoab | 0.689 | cirbpb | -0.580 |
| atp2a1 | 0.681 | khdrbs1a | -0.575 |
| ak1 | 0.676 | hnrnpaba | -0.575 |
| actn3a | 0.669 | hmgb2b | -0.567 |
| ldb3b | 0.666 | ran | -0.565 |
| neb | 0.665 | cirbpa | -0.557 |
| nme2b.2 | 0.664 | hmga1a | -0.552 |
| ank1a | 0.653 | si:ch211-222l21.1 | -0.545 |
| myom1a | 0.650 | si:ch73-281n10.2 | -0.543 |
| smyd1a | 0.648 | hnrnpabb | -0.542 |
| actc1b | 0.647 | hmgb2a | -0.541 |
| casq2 | 0.646 | ptmab | -0.530 |
| pkmb | 0.645 | hnrnpa0b | -0.519 |
| tpi1b | 0.643 | setb | -0.518 |
| actn3b | 0.642 | hmgn2 | -0.514 |
| prx | 0.641 | syncrip | -0.514 |
| casq1b | 0.629 | cbx3a | -0.511 |
| si:ch211-266g18.10 | 0.625 | eef1a1l1 | -0.509 |
| si:ch73-367p23.2 | 0.625 | ubc | -0.508 |
| tmem38a | 0.623 | hmgn7 | -0.505 |
| pygma | 0.621 | fthl27 | -0.502 |
| tnnc2 | 0.619 | naca | -0.502 |
| tmod4 | 0.618 | snrpf | -0.501 |
| srl | 0.614 | hmgn6 | -0.494 |
| ttn.2 | 0.606 | snrpb | -0.485 |
| myom1b | 0.605 | nucks1a | -0.484 |
| XLOC-025819 | 0.605 | anp32b | -0.482 |
| eno1a | 0.602 | tuba8l4 | -0.477 |
| ckmt2b | 0.601 | snrpd1 | -0.474 |
| CABZ01078594.1 | 0.599 | seta | -0.474 |