post-GPI attachment to proteins phospholipase 3
ZFIN 
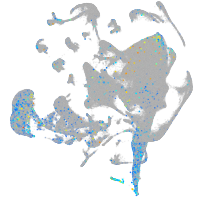
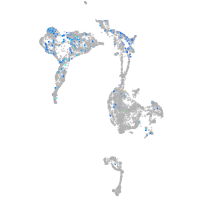
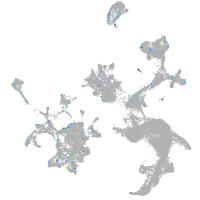
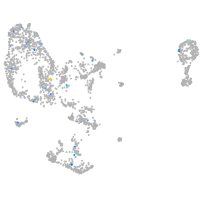
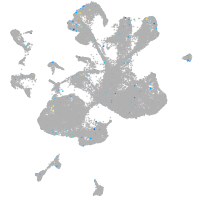
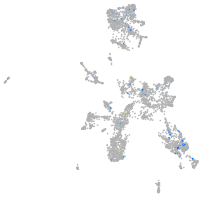
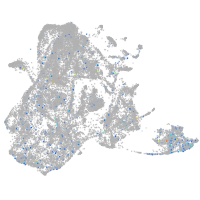
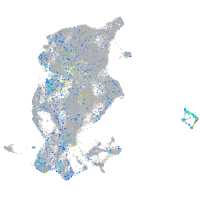
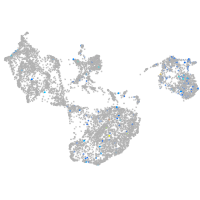
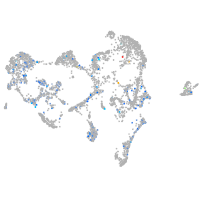
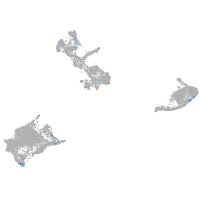
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC797168 | 0.111 | h3f3a | -0.040 |
| cbln2a | 0.092 | ptmab | -0.039 |
| XLOC-028486 | 0.089 | hnrnpa0l | -0.039 |
| ngfa | 0.077 | rps10 | -0.039 |
| BX572077.1 | 0.076 | fabp7a | -0.036 |
| sox17 | 0.073 | hmgn6 | -0.033 |
| LOC101887065 | 0.072 | slc25a5 | -0.033 |
| LOC110438228 | 0.071 | rpl37 | -0.032 |
| zte38 | 0.070 | hmgn2 | -0.031 |
| BX649490.4 | 0.066 | zgc:114188 | -0.030 |
| trpm2 | 0.065 | h2afva | -0.030 |
| BX005108.2 | 0.064 | fabp3 | -0.029 |
| aifm5 | 0.064 | zgc:153409 | -0.029 |
| capn3a | 0.063 | rps17 | -0.028 |
| crygm2d11 | 0.063 | h2afvb | -0.028 |
| si:ch211-255g12.6 | 0.062 | ptmaa | -0.027 |
| crygm2d4 | 0.062 | crx | -0.026 |
| crygm2d10 | 0.061 | neurod4 | -0.026 |
| XLOC-020173 | 0.060 | hmgb2a | -0.025 |
| fndc5a | 0.060 | si:ch73-1a9.3 | -0.025 |
| crygm2d8 | 0.059 | ppiab | -0.025 |
| endou2 | 0.058 | alyref | -0.025 |
| lctla | 0.058 | cct2 | -0.025 |
| XLOC-020175 | 0.058 | hsp90ab1 | -0.025 |
| crygm2d5 | 0.058 | sinhcafl | -0.024 |
| crygm2d13 | 0.058 | cox5aa | -0.024 |
| BEND4 | 0.058 | cadm3 | -0.024 |
| BFSP1 | 0.058 | atp5pd | -0.023 |
| muc5d | 0.058 | ubc | -0.023 |
| cryba1l2 | 0.057 | elocb | -0.023 |
| si:ch211-152c2.3 | 0.057 | rps7 | -0.023 |
| crygm2d1 | 0.057 | ndrg1b | -0.023 |
| crygm2d18 | 0.057 | rps27.1 | -0.022 |
| LOC110438555 | 0.056 | si:ch1073-429i10.3.1 | -0.022 |
| crygm2d7 | 0.056 | foxg1b | -0.022 |