penta-EF-hand domain containing 1
ZFIN 
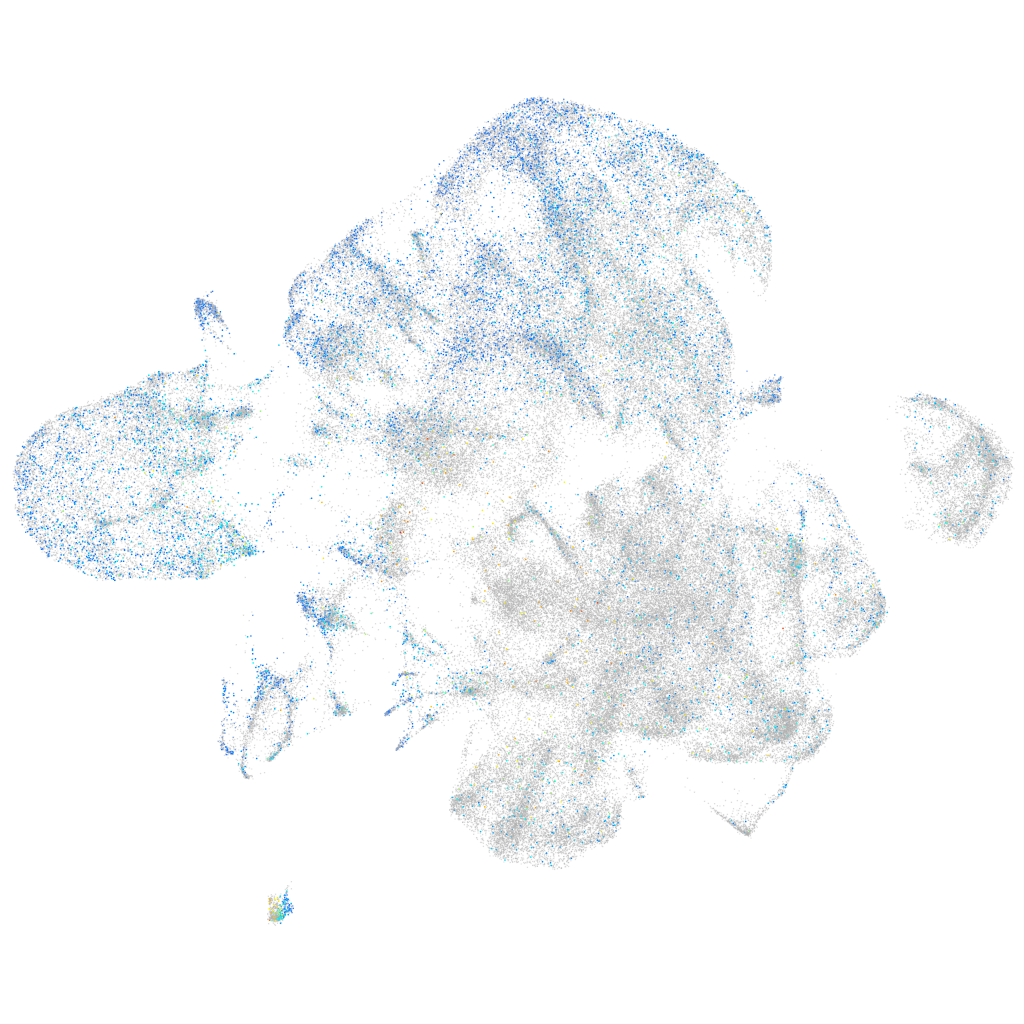
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb2a | 0.117 | rtn1a | -0.108 |
| anp32b | 0.110 | gpm6aa | -0.105 |
| id1 | 0.108 | stmn1b | -0.104 |
| pcna | 0.107 | elavl3 | -0.100 |
| banf1 | 0.106 | tuba1c | -0.100 |
| chaf1a | 0.104 | gpm6ab | -0.099 |
| npm1a | 0.103 | ckbb | -0.094 |
| nop58 | 0.102 | ptmaa | -0.093 |
| lig1 | 0.100 | gng3 | -0.092 |
| tuba8l4 | 0.099 | rtn1b | -0.090 |
| ranbp1 | 0.098 | sncb | -0.088 |
| fbl | 0.097 | elavl4 | -0.087 |
| selenoh | 0.096 | atp6v0cb | -0.085 |
| nop56 | 0.095 | tmsb | -0.083 |
| dkc1 | 0.095 | nova2 | -0.081 |
| snu13b | 0.095 | rnasekb | -0.078 |
| vamp3 | 0.094 | si:dkeyp-75h12.5 | -0.078 |
| hspb1 | 0.094 | stx1b | -0.078 |
| si:ch73-281n10.2 | 0.093 | zc4h2 | -0.076 |
| rrm1 | 0.092 | tubb5 | -0.076 |
| ccng1 | 0.092 | fez1 | -0.075 |
| eif5a2 | 0.092 | vamp2 | -0.075 |
| nasp | 0.091 | COX3 | -0.075 |
| zgc:110425 | 0.091 | stxbp1a | -0.073 |
| hmga1a | 0.091 | ywhag2 | -0.073 |
| akap12b | 0.091 | aplp1 | -0.073 |
| fen1 | 0.090 | celf2 | -0.071 |
| rpa3 | 0.090 | stmn2a | -0.071 |
| sdc4 | 0.089 | kdm6bb | -0.071 |
| nhp2 | 0.088 | nsg2 | -0.071 |
| eef1a1l1 | 0.088 | gng2 | -0.071 |
| cnbpa | 0.087 | ywhah | -0.070 |
| dek | 0.087 | id4 | -0.070 |
| snrpb | 0.087 | gap43 | -0.070 |
| mibp | 0.087 | dpysl3 | -0.069 |