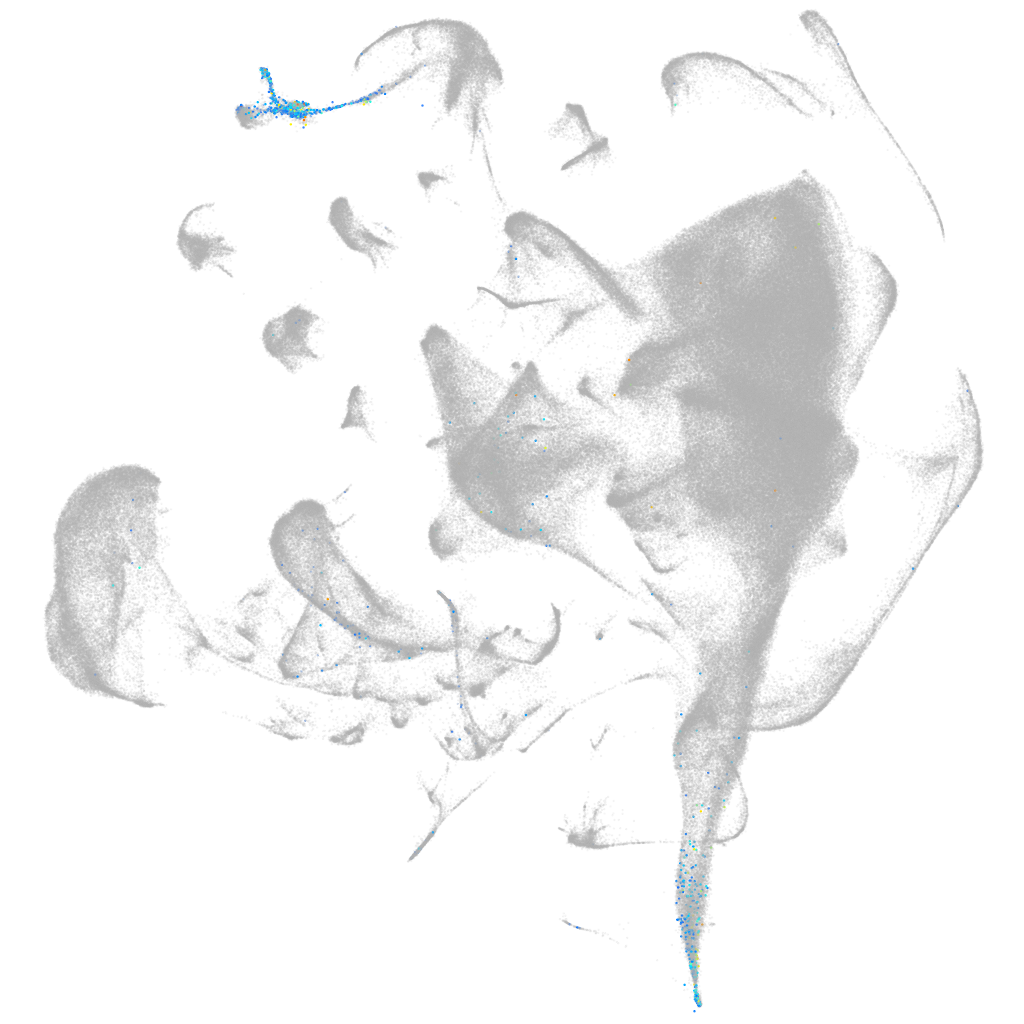
PEAK family member 3
ZFIN 
Other cell groups


all cells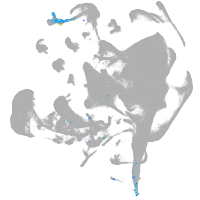 |
blastomeres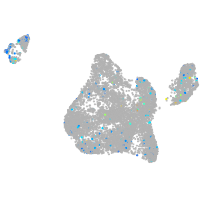 |
PGCs |
endoderm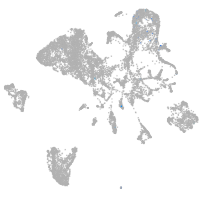 |
axial mesoderm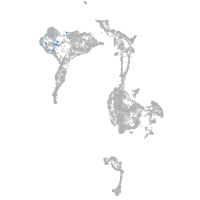 |
muscle  |
mural cells / non-skeletal muscle 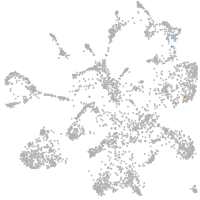 |
mesenchyme  |
hematopoietic / vasculature 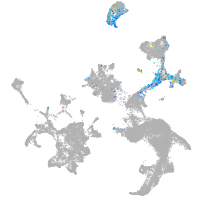 |
pronephros 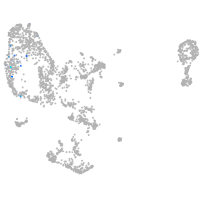 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-214p16.1 | 0.305 | tuba1c | -0.040 |
| lpar5a | 0.297 | hmgb3a | -0.039 |
| si:dkey-185m8.2 | 0.288 | nova2 | -0.039 |
| si:ch73-208g10.1 | 0.273 | mdka | -0.037 |
| wasa | 0.273 | tuba1a | -0.036 |
| tnfaip8l2a | 0.272 | mdkb | -0.035 |
| laptm5 | 0.270 | fabp3 | -0.034 |
| grap2b | 0.269 | gpm6aa | -0.033 |
| itgae.2 | 0.266 | tmeff1b | -0.033 |
| coro1a | 0.260 | her15.1 | -0.033 |
| wasb | 0.255 | si:ch211-137a8.4 | -0.032 |
| plek | 0.251 | elavl3 | -0.031 |
| rasal3 | 0.247 | hmgb1b | -0.031 |
| vaspa | 0.247 | rtn1a | -0.031 |
| hmha1b | 0.245 | marcksl1a | -0.031 |
| mir223 | 0.244 | fam168a | -0.030 |
| parvg | 0.241 | h1f0 | -0.030 |
| unc13d | 0.240 | rnasekb | -0.029 |
| si:rp71-68n21.9 | 0.237 | XLOC-003690 | -0.028 |
| ccr9a | 0.235 | si:ch1073-429i10.3.1 | -0.028 |
| FERMT3 (1 of many) | 0.232 | gpm6ab | -0.027 |
| glipr1a | 0.229 | stmn1b | -0.027 |
| cd7al | 0.226 | tmsb | -0.027 |
| fmnl1a | 0.223 | tubb5 | -0.027 |
| csf1rb | 0.221 | foxp4 | -0.026 |
| lcp1 | 0.221 | si:ch73-46j18.5 | -0.026 |
| ptprc | 0.221 | sparc | -0.026 |
| rgs13 | 0.221 | XLOC-003689 | -0.026 |
| si:ch73-248e21.5 | 0.220 | mbd3b | -0.026 |
| ccdc88b | 0.218 | angptl4 | -0.025 |
| nrros | 0.215 | cdh2 | -0.025 |
| si:dkeyp-68b7.12 | 0.215 | cldn5a | -0.025 |
| CU019662.1 | 0.214 | fabp7a | -0.025 |
| myo1f | 0.214 | meis1b | -0.025 |
| grap2a | 0.212 | msi1 | -0.025 |