PDZ domain containing 3b
ZFIN 
Other cell groups


all cells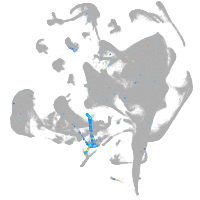 |
blastomeres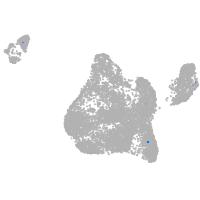 |
PGCs |
endoderm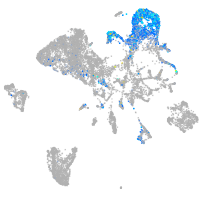 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 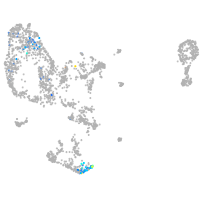 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX649516.4 | 0.773 | rpl22 | -0.054 |
| XLOC-002703 | 0.697 | rps8a | -0.047 |
| zp3.2 | 0.657 | rps26 | -0.046 |
| XLOC-012358 | 0.586 | mt-co1 | -0.044 |
| XLOC-016096 | 0.582 | btf3 | -0.044 |
| CR387932.1 | 0.522 | eef1g | -0.040 |
| acot7 | 0.482 | ptmaa | -0.039 |
| aldh8a1 | 0.475 | fthl27 | -0.036 |
| XLOC-044090 | 0.437 | mt-nd1 | -0.034 |
| CHST13 | 0.424 | atp5mc3b | -0.034 |
| sult3st1 | 0.415 | hmgn2 | -0.033 |
| cnfn | 0.342 | hmgn7 | -0.033 |
| XLOC-032836 | 0.331 | cirbpa | -0.033 |
| cldnc | 0.325 | cox6a1 | -0.033 |
| si:dkey-69o16.5 | 0.307 | h3f3c | -0.033 |
| emid1 | 0.306 | COX5B | -0.032 |
| srl | 0.300 | cox8a | -0.031 |
| stard8 | 0.300 | cfl1 | -0.031 |
| ptrh1 | 0.296 | hnrnpaba | -0.031 |
| chia.2 | 0.282 | rps17 | -0.031 |
| rgs2 | 0.275 | hnrnpa0l | -0.030 |
| dhrs9 | 0.274 | cox7c | -0.030 |
| LOC101883323 | 0.266 | sec61g | -0.030 |
| mmp15a | 0.250 | slc25a3b | -0.030 |
| si:ch211-196g2.4 | 0.208 | zgc:193541 | -0.030 |
| nadk2 | 0.205 | atp5f1b | -0.029 |
| oplah | 0.196 | eif3f | -0.029 |
| tmem201 | 0.189 | oaz1b | -0.028 |
| tas1r2.2 | 0.181 | actb2 | -0.028 |
| rrp7a | 0.181 | calm2b | -0.027 |
| suclg2 | 0.181 | tubb4b | -0.027 |
| chaf1b | 0.180 | dynll1 | -0.027 |
| FP101887.1 | 0.179 | rpl3 | -0.027 |
| unm-hu7910 | 0.179 | marcksl1a | -0.026 |
| nub1 | 0.175 | tubb2b | -0.026 |




