"pyridoxal (pyridoxine, vitamin B6) kinase a"
ZFIN 
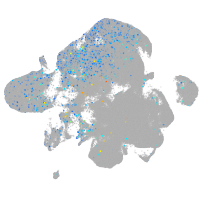
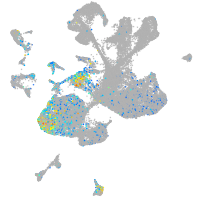
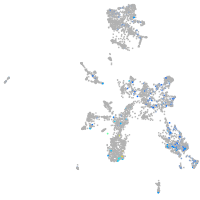
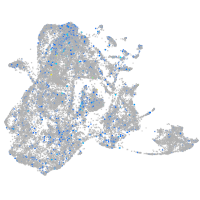
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43 | 0.237 | elavl3 | -0.078 |
| hepacama | 0.233 | stmn1b | -0.069 |
| si:ch211-66e2.5 | 0.229 | tubb5 | -0.069 |
| hspb15 | 0.221 | gng3 | -0.064 |
| slc4a4a | 0.215 | sncb | -0.063 |
| gpr37l1b | 0.214 | myt1b | -0.060 |
| slc6a11b | 0.213 | rtn1a | -0.059 |
| atp1a1b | 0.207 | stx1b | -0.058 |
| slc3a2a | 0.204 | vamp2 | -0.058 |
| atp1b4 | 0.196 | zc4h2 | -0.058 |
| ptn | 0.190 | chd4a | -0.057 |
| mfge8a | 0.181 | elavl4 | -0.056 |
| glula | 0.178 | si:dkey-276j7.1 | -0.056 |
| ppap2d | 0.176 | myt1a | -0.055 |
| cldn7a | 0.173 | tmsb | -0.055 |
| slc1a3b | 0.173 | stxbp1a | -0.055 |
| si:dkey-245n4.2 | 0.167 | atp6v0cb | -0.054 |
| slc1a2b | 0.164 | hmgb3a | -0.054 |
| FO704813.1 | 0.161 | marcksb | -0.054 |
| mlc1 | 0.161 | stmn2a | -0.054 |
| si:ch1073-303k11.2 | 0.157 | nsg2 | -0.053 |
| si:ch211-180a12.2 | 0.155 | ywhag2 | -0.053 |
| ptgdsb.2 | 0.154 | gap43 | -0.051 |
| abi3a | 0.152 | hnrnpa0a | -0.051 |
| qki2 | 0.150 | scrt2 | -0.051 |
| eno1b | 0.148 | tuba1c | -0.051 |
| efhd1 | 0.147 | cnrip1a | -0.050 |
| endouc | 0.147 | epb41a | -0.050 |
| sept8b | 0.145 | rtn1b | -0.050 |
| fjx1 | 0.144 | atp1a3a | -0.049 |
| pald1b | 0.140 | snap25a | -0.049 |
| slc7a10b | 0.139 | aplp1 | -0.048 |
| anxa13 | 0.138 | gpm6ab | -0.048 |
| ca4a | 0.135 | ywhah | -0.048 |
| cdo1 | 0.135 | jagn1a | -0.047 |