proprotein convertase subtilisin/kexin type 6
ZFIN 
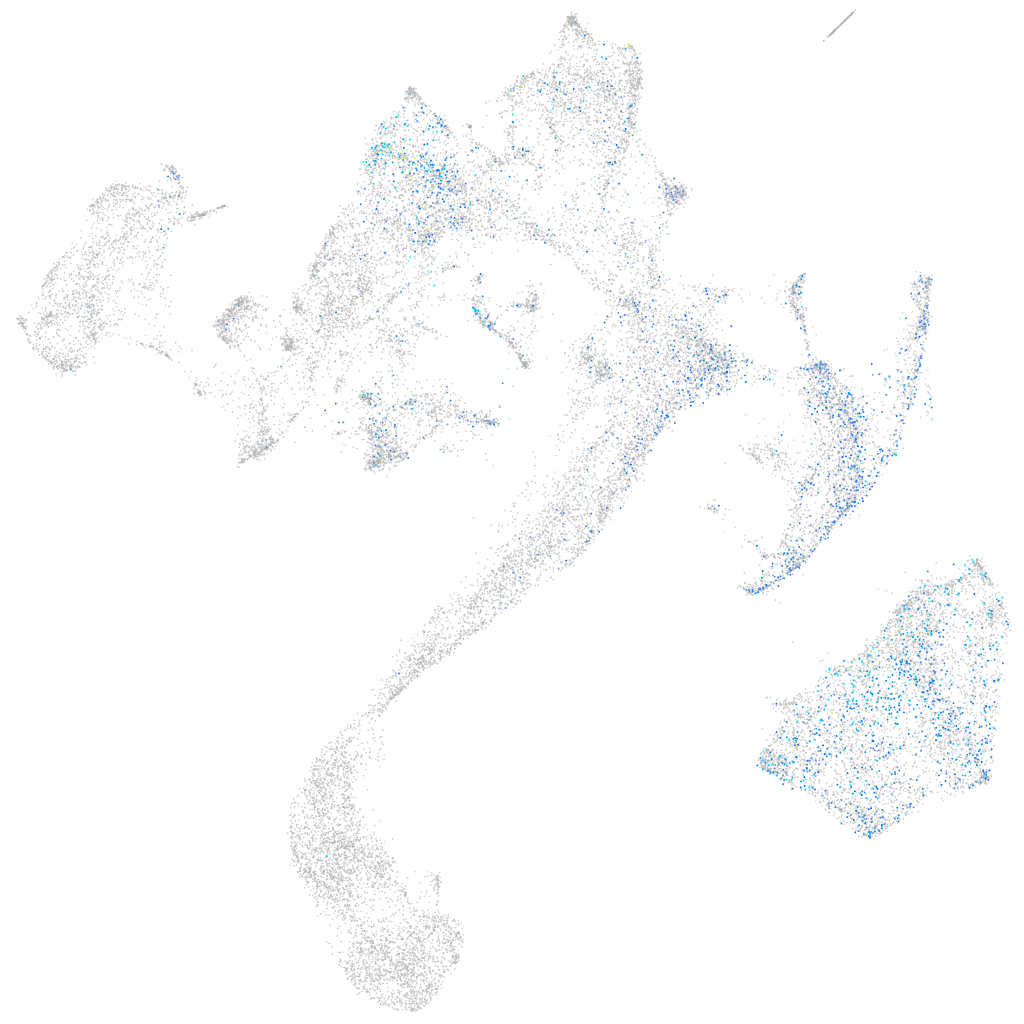
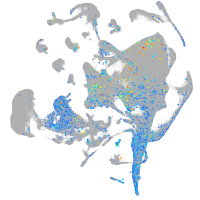
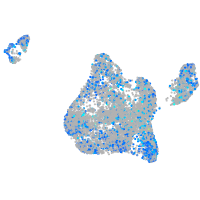
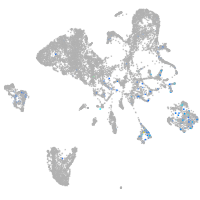
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.205 | actc1b | -0.168 |
| cdx4 | 0.189 | rpl37 | -0.165 |
| anp32e | 0.187 | atp2a1 | -0.153 |
| acin1a | 0.183 | ak1 | -0.153 |
| hes6 | 0.182 | ckmb | -0.153 |
| nucks1a | 0.180 | zgc:114188 | -0.152 |
| hnrnpub | 0.179 | ckma | -0.151 |
| marcksb | 0.178 | pabpc4 | -0.151 |
| si:ch73-1a9.3 | 0.177 | ttn.2 | -0.149 |
| NC-002333.4 | 0.174 | tmem38a | -0.149 |
| tbx16 | 0.174 | rps10 | -0.149 |
| rbm38 | 0.173 | aldoab | -0.146 |
| pnrc2 | 0.173 | rps17 | -0.143 |
| ptmab | 0.173 | tnnc2 | -0.138 |
| hnrnpa1b | 0.173 | mylpfa | -0.138 |
| seta | 0.171 | acta1b | -0.136 |
| smarca4a | 0.171 | neb | -0.135 |
| anp32a | 0.171 | gamt | -0.135 |
| srrm1 | 0.169 | ldb3b | -0.135 |
| smc1al | 0.169 | atp5if1b | -0.135 |
| mki67 | 0.169 | eno1a | -0.134 |
| ctnnb1 | 0.168 | myl1 | -0.134 |
| ilf3b | 0.168 | idh2 | -0.134 |
| hnrnpa1a | 0.168 | eno3 | -0.134 |
| hmga1a | 0.166 | atp5f1b | -0.133 |
| snrnp70 | 0.165 | srl | -0.133 |
| cdx1a | 0.164 | ttn.1 | -0.133 |
| syncrip | 0.164 | atp5meb | -0.133 |
| wnt5b | 0.164 | atp5mc3b | -0.133 |
| safb | 0.164 | vdac2 | -0.132 |
| cirbpa | 0.163 | CABZ01078594.1 | -0.131 |
| hnrnpm | 0.163 | tnnt3a | -0.131 |
| si:ch73-281n10.2 | 0.163 | suclg1 | -0.131 |
| top1l | 0.161 | cav3 | -0.131 |
| rbm4.3 | 0.161 | si:ch73-367p23.2 | -0.130 |