proprotein convertase subtilisin/kexin type 6
ZFIN 
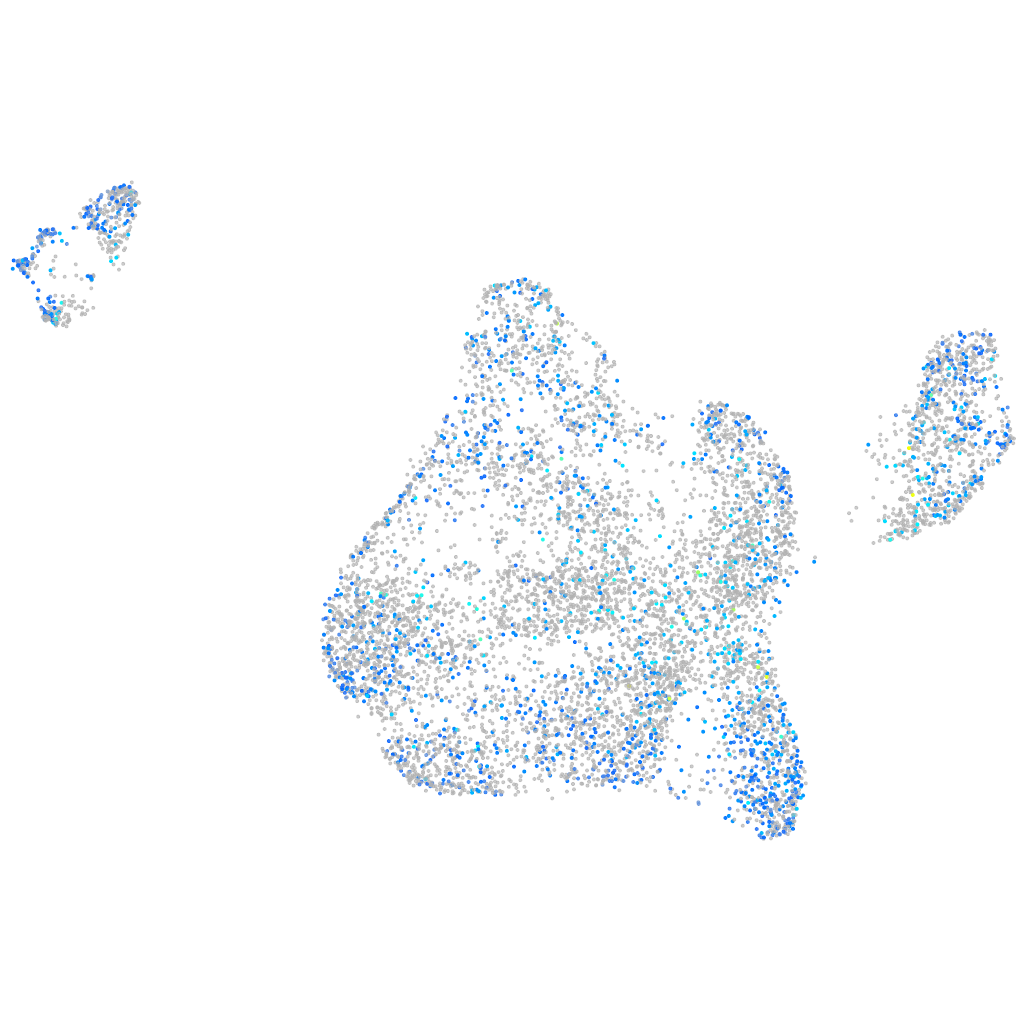
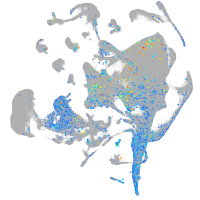
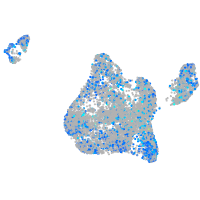
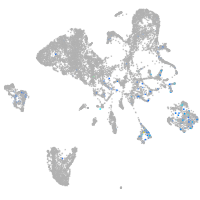
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gkap1 | 0.086 | NC-002333.4 | -0.061 |
| sh3pxd2b | 0.081 | hspb1 | -0.055 |
| zp3.2 | 0.078 | hnrnpa0b | -0.051 |
| zc3h13 | 0.077 | mt-cyb | -0.051 |
| atp2b1a | 0.076 | mt-co2 | -0.050 |
| rif1 | 0.075 | polr3gla | -0.048 |
| h1m | 0.073 | slc25a5 | -0.043 |
| cacul1 | 0.071 | ube2e2 | -0.043 |
| dgcr8 | 0.071 | rpl7 | -0.041 |
| rbm26 | 0.069 | hnrnpa0a | -0.040 |
| ehmt1b | 0.069 | si:ch211-222l21.1 | -0.039 |
| ktn1 | 0.069 | rplp0 | -0.039 |
| EML4 | 0.068 | si:ch211-152c2.3 | -0.038 |
| wasf2 | 0.068 | mid1ip1b | -0.037 |
| ccng1 | 0.068 | apoeb | -0.037 |
| XLOC-035411 | 0.067 | ybx1 | -0.037 |
| thoc2 | 0.067 | ppiab | -0.036 |
| prkrip1 | 0.067 | ptmab | -0.036 |
| aif1l | 0.066 | rpl6 | -0.036 |
| lamp2 | 0.066 | ubb | -0.036 |
| uhrf1 | 0.066 | hmgn2 | -0.035 |
| lgmn | 0.065 | tma7 | -0.035 |
| ppp1r9ala | 0.065 | COX3 | -0.035 |
| pnrc2 | 0.065 | snrpb | -0.035 |
| pttg1ipb | 0.065 | crtap | -0.034 |
| cmtm6 | 0.064 | hmgb2b | -0.033 |
| usp10 | 0.064 | xk | -0.032 |
| stx4 | 0.064 | bmp4 | -0.032 |
| brd8 | 0.064 | aldob | -0.031 |
| supt6h | 0.063 | rps4x | -0.031 |
| zfand5b | 0.063 | marcksl1b | -0.031 |
| tpm2 | 0.063 | mrpl54 | -0.031 |
| nanog | 0.062 | rps15 | -0.030 |
| gnaia | 0.062 | h2afvb | -0.030 |
| gclc | 0.062 | tuba8l4 | -0.030 |