polycomb group ring finger 1
ZFIN 
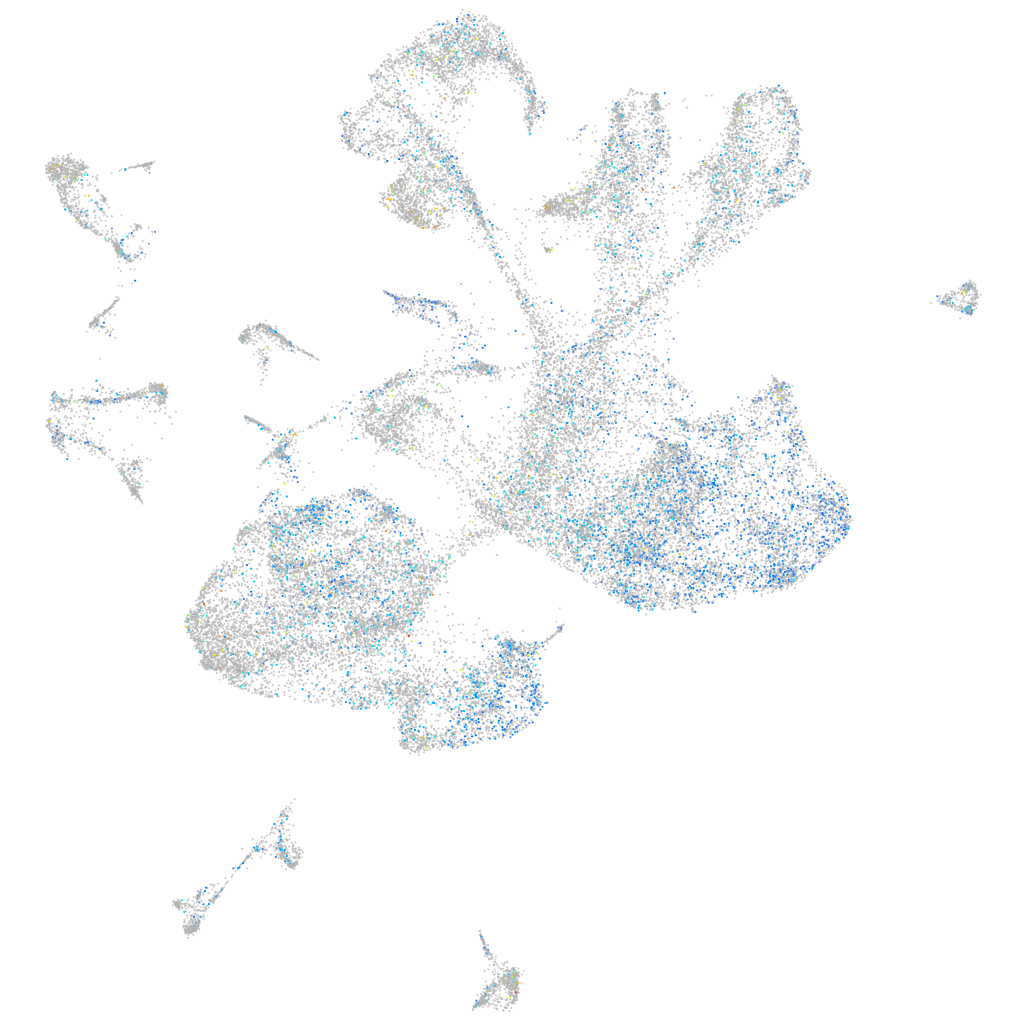
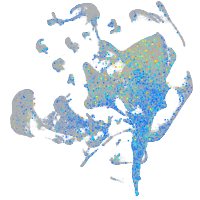
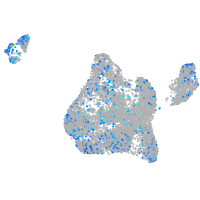
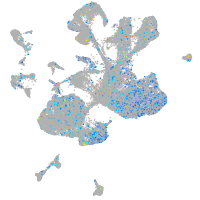
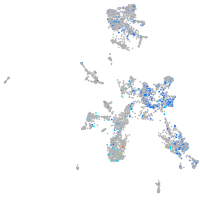
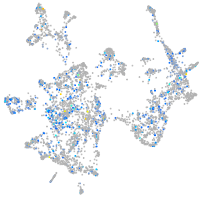
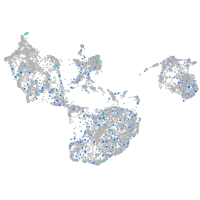
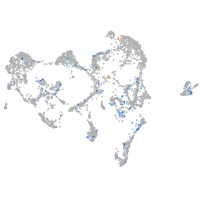
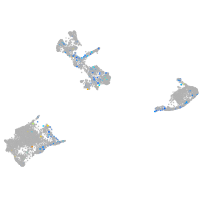
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.115 | ckbb | -0.050 |
| mcm5 | 0.114 | slc1a2b | -0.046 |
| mcm7 | 0.114 | syt11a | -0.044 |
| nasp | 0.113 | slc4a4a | -0.042 |
| hells | 0.112 | gapdhs | -0.042 |
| cdca7a | 0.112 | rnasekb | -0.042 |
| mcm6 | 0.110 | cx43 | -0.041 |
| mcm4 | 0.109 | rtn1a | -0.041 |
| mcm2 | 0.106 | ptn | -0.041 |
| mcm3 | 0.104 | aldocb | -0.041 |
| slbp | 0.104 | ndrg3a | -0.040 |
| fen1 | 0.102 | anxa13 | -0.039 |
| stmn1a | 0.101 | rpz5 | -0.038 |
| rpa3 | 0.100 | onecut1 | -0.037 |
| ranbp1 | 0.099 | si:dkeyp-75h12.5 | -0.037 |
| npm1a | 0.099 | calm1a | -0.037 |
| chaf1a | 0.099 | elavl3 | -0.037 |
| gmnn | 0.098 | aplp1 | -0.037 |
| nop58 | 0.098 | gpm6ab | -0.036 |
| dut | 0.097 | gabarapa | -0.036 |
| rpa2 | 0.097 | atp6v0cb | -0.036 |
| snrpd1 | 0.096 | smox | -0.036 |
| selenoh | 0.094 | hepacama | -0.035 |
| anp32b | 0.093 | acbd7 | -0.035 |
| hmgb2b | 0.093 | proza | -0.035 |
| rrm1 | 0.092 | efhd1 | -0.035 |
| dkc1 | 0.092 | myt1b | -0.035 |
| hmgb2a | 0.092 | IGLON5 | -0.034 |
| cdca7b | 0.091 | pvalb1 | -0.034 |
| tuba8l4 | 0.091 | nrcama | -0.034 |
| snu13b | 0.091 | slc6a1b | -0.034 |
| lig1 | 0.091 | igfbp7 | -0.034 |
| setb | 0.090 | sept8b | -0.034 |
| her2 | 0.090 | syt2a | -0.034 |
| ran | 0.090 | cldn7a | -0.034 |