polybromo 1
ZFIN 
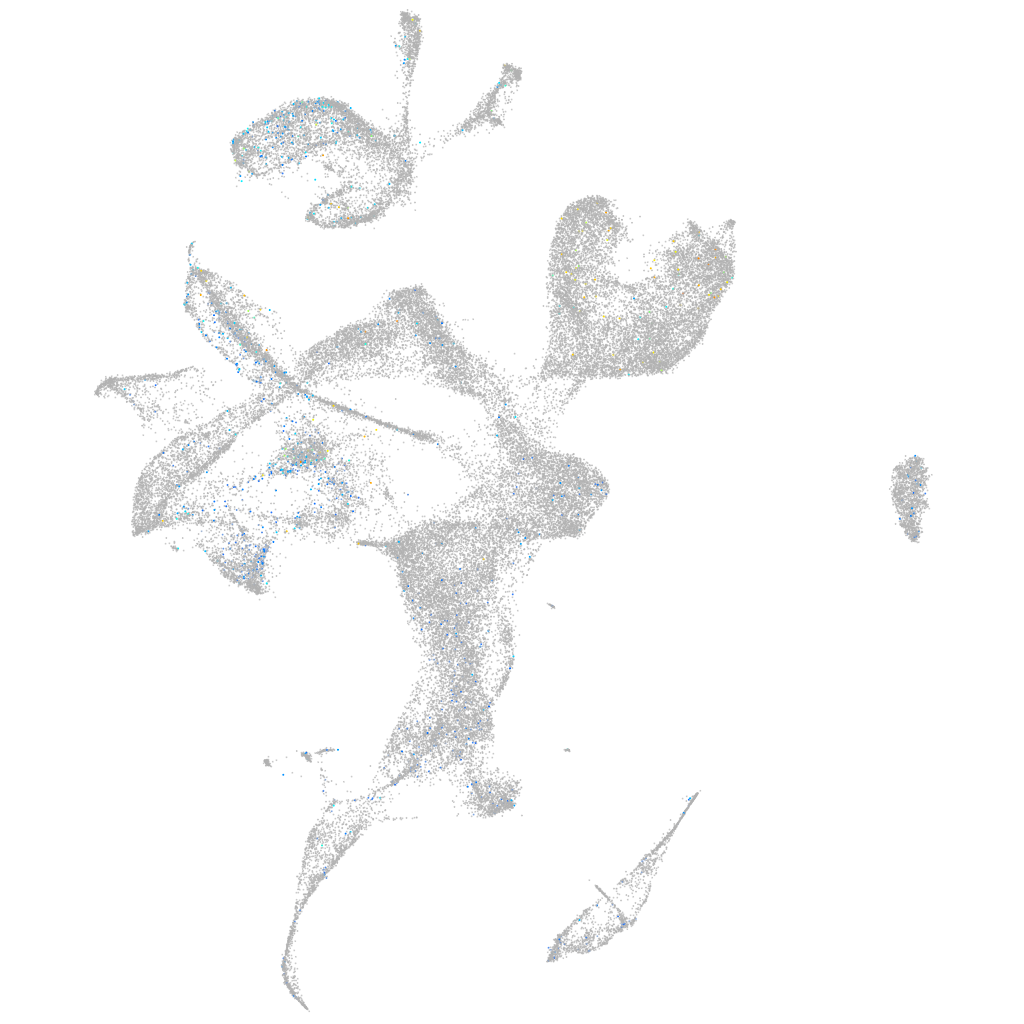
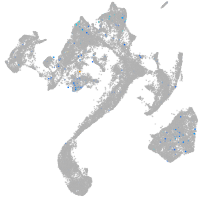
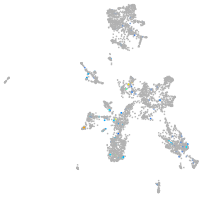
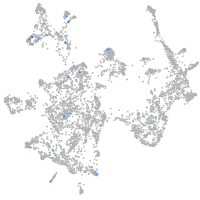
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.085 | hmgb2a | -0.055 |
| stmn2a | 0.083 | mdka | -0.036 |
| elavl3 | 0.083 | fabp7a | -0.034 |
| stx1b | 0.083 | ahcy | -0.033 |
| rtn1b | 0.082 | msi1 | -0.030 |
| elavl4 | 0.081 | pcna | -0.030 |
| celf5a | 0.081 | hmga1a | -0.030 |
| gng3 | 0.080 | dek | -0.029 |
| ywhag2 | 0.079 | hmgb2b | -0.029 |
| stmn1b | 0.077 | rrm1 | -0.029 |
| zgc:65894 | 0.076 | nutf2l | -0.028 |
| syt1a | 0.076 | her15.1 | -0.027 |
| sv2a | 0.076 | rrm2 | -0.027 |
| sncb | 0.075 | mibp | -0.026 |
| ppp1r14ba | 0.072 | lbr | -0.026 |
| myt1la | 0.071 | stmn1a | -0.026 |
| zgc:153426 | 0.071 | neurod4 | -0.025 |
| snap25a | 0.070 | ncapg | -0.025 |
| ppfia3 | 0.069 | hes2.2 | -0.025 |
| zc4h2 | 0.069 | si:dkey-151g10.6 | -0.025 |
| maptb | 0.067 | cks1b | -0.025 |
| map1aa | 0.067 | rplp2l | -0.025 |
| pbx3b | 0.067 | chaf1a | -0.025 |
| LOC797168 | 0.067 | zgc:110540 | -0.025 |
| eno2 | 0.066 | ccna2 | -0.025 |
| sprn | 0.066 | im:7152348 | -0.024 |
| slc32a1 | 0.066 | rpl8 | -0.024 |
| fez1 | 0.065 | ccnd1 | -0.024 |
| gabrb4 | 0.065 | rps7 | -0.024 |
| syn2a | 0.065 | CABZ01005379.1 | -0.024 |
| rtn1a | 0.065 | dut | -0.024 |
| vsnl1b | 0.064 | rps29 | -0.024 |
| celf4 | 0.064 | rps20 | -0.024 |
| ebf3a | 0.064 | rps25 | -0.024 |
| BX005003.1 | 0.064 | tspan7 | -0.024 |