paired box 6b
ZFIN 
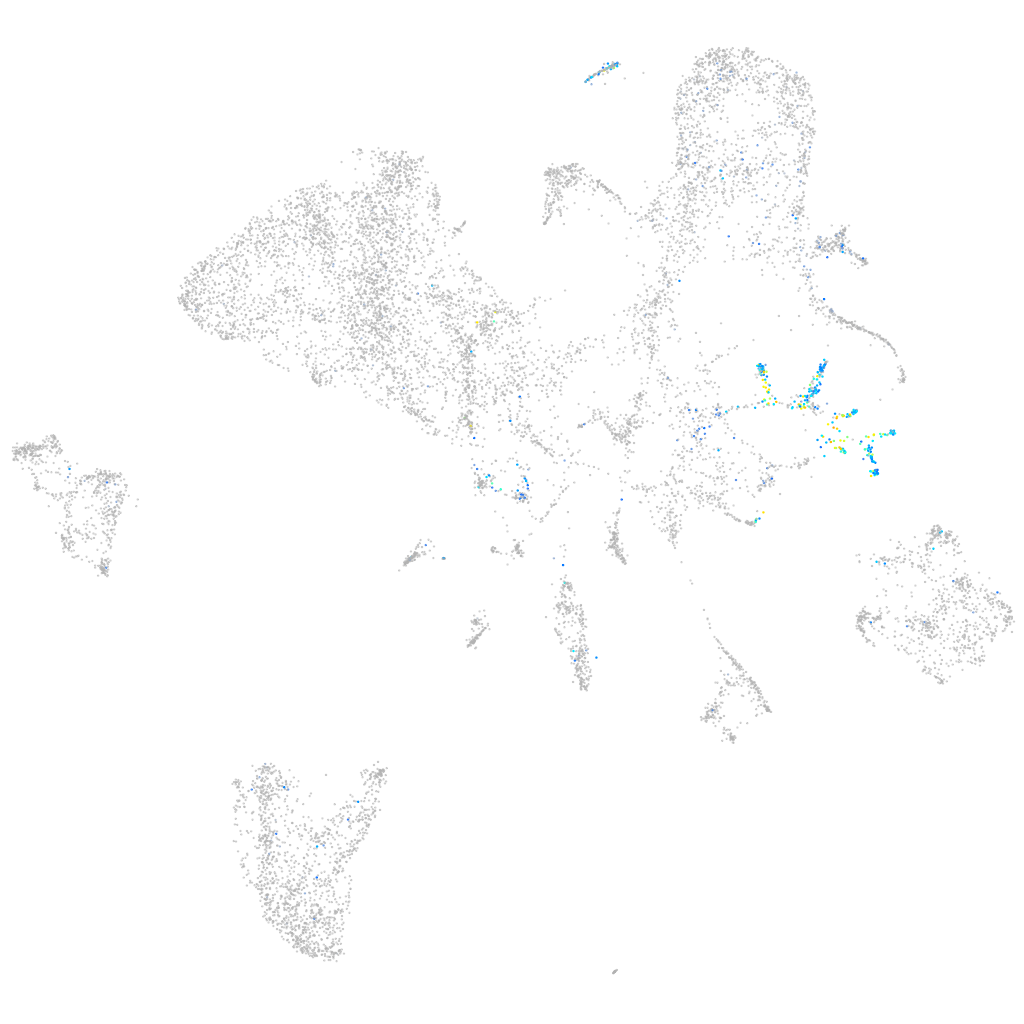
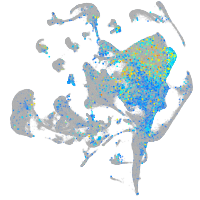
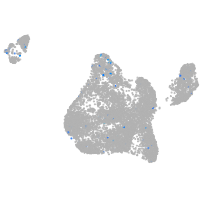
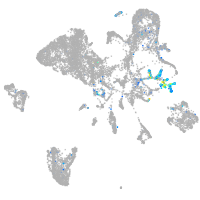
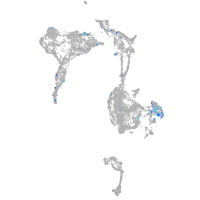
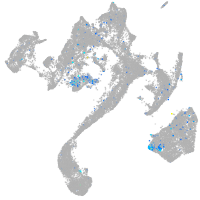
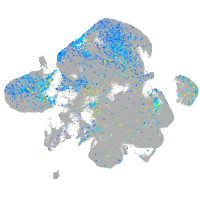
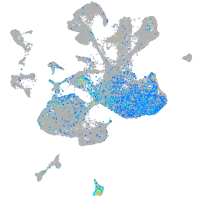
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.656 | ahcy | -0.188 |
| neurod1 | 0.622 | gapdh | -0.184 |
| scgn | 0.594 | gamt | -0.172 |
| mir7a-1 | 0.536 | aldob | -0.157 |
| rprmb | 0.534 | hdlbpa | -0.154 |
| pcsk1nl | 0.533 | gatm | -0.153 |
| si:dkey-153k10.9 | 0.531 | bhmt | -0.147 |
| kcnj11 | 0.529 | gstr | -0.147 |
| isl1 | 0.523 | mat1a | -0.145 |
| insm1a | 0.509 | nupr1b | -0.144 |
| vat1 | 0.470 | fbp1b | -0.143 |
| syt1a | 0.469 | aldh6a1 | -0.143 |
| pcsk1 | 0.467 | cx32.3 | -0.142 |
| tspan7b | 0.463 | gstp1 | -0.141 |
| mir375-2 | 0.461 | apoa1b | -0.141 |
| rasd4 | 0.455 | apoa4b.1 | -0.140 |
| ndufa4l2a | 0.446 | fabp3 | -0.139 |
| fev | 0.437 | glud1b | -0.139 |
| scg2a | 0.433 | cebpd | -0.137 |
| lysmd2 | 0.432 | gstt1a | -0.137 |
| cacna1c | 0.431 | si:dkey-16p21.8 | -0.132 |
| nkx2.2a | 0.431 | agxta | -0.132 |
| c2cd4a | 0.430 | apoc2 | -0.131 |
| pcsk2 | 0.427 | gnmt | -0.130 |
| myt1b | 0.426 | aldh7a1 | -0.130 |
| gpc1a | 0.421 | eno3 | -0.129 |
| vamp2 | 0.418 | eef1da | -0.128 |
| pygma | 0.415 | mgst1.2 | -0.128 |
| rims2a | 0.411 | ppa1b | -0.128 |
| vwa5b2 | 0.408 | agxtb | -0.127 |
| foxo1a | 0.402 | aqp12 | -0.126 |
| camk1da | 0.400 | apoc1 | -0.125 |
| aldocb | 0.397 | apoa2 | -0.125 |
| SLC5A10 | 0.396 | afp4 | -0.125 |
| abhd15a | 0.396 | tfa | -0.122 |