PAS domain containing serine/threonine kinase
ZFIN 
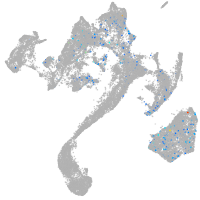
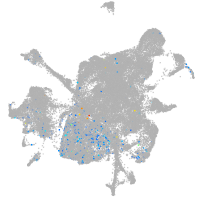
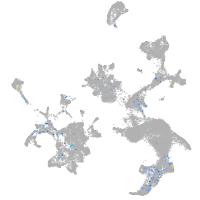
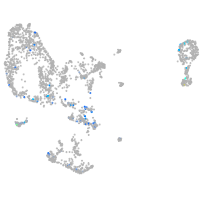
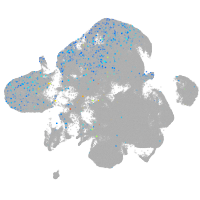
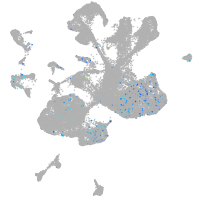
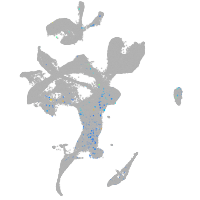
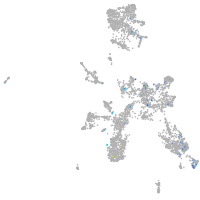
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.124 | actc1b | -0.082 |
| stmn1a | 0.123 | ttn.1 | -0.078 |
| zgc:110540 | 0.122 | aldoab | -0.076 |
| lbr | 0.119 | ttn.2 | -0.076 |
| tubb2b | 0.118 | tmem38a | -0.075 |
| chaf1a | 0.118 | ak1 | -0.074 |
| tuba8l4 | 0.117 | ckma | -0.073 |
| rrm1 | 0.117 | atp2a1 | -0.073 |
| rrm2 | 0.115 | mybphb | -0.072 |
| mibp | 0.114 | tnnc2 | -0.071 |
| cks1b | 0.113 | ckmb | -0.071 |
| rbbp4 | 0.112 | klhl41b | -0.070 |
| CABZ01005379.1 | 0.110 | desma | -0.070 |
| dek | 0.109 | acta1b | -0.069 |
| rpa3 | 0.109 | pabpc4 | -0.069 |
| nasp | 0.107 | neb | -0.068 |
| sumo3b | 0.107 | srl | -0.068 |
| si:ch211-222l21.1 | 0.107 | ldb3a | -0.067 |
| fen1 | 0.107 | CABZ01078594.1 | -0.067 |
| hmgb2a | 0.107 | tpma | -0.066 |
| rpa2 | 0.106 | cav3 | -0.066 |
| asf1ba | 0.103 | gapdh | -0.066 |
| slbp | 0.102 | mylpfa | -0.066 |
| banf1 | 0.102 | actc1a | -0.066 |
| fbxo5 | 0.101 | acta1a | -0.065 |
| ccna2 | 0.100 | gamt | -0.065 |
| mki67 | 0.100 | eno3 | -0.065 |
| seta | 0.100 | vdac3 | -0.065 |
| zgc:165555.3 | 0.100 | ldb3b | -0.064 |
| hmgb2b | 0.100 | myl1 | -0.064 |
| smc2 | 0.100 | actn3b | -0.063 |
| dhfr | 0.099 | nme2b.2 | -0.063 |
| hmgb1b | 0.099 | eno1a | -0.063 |
| si:ch211-288g17.3 | 0.099 | zgc:101853 | -0.062 |
| zgc:110216 | 0.098 | si:ch73-367p23.2 | -0.062 |