PAXIP1 associated glutamate-rich protein 1
ZFIN 
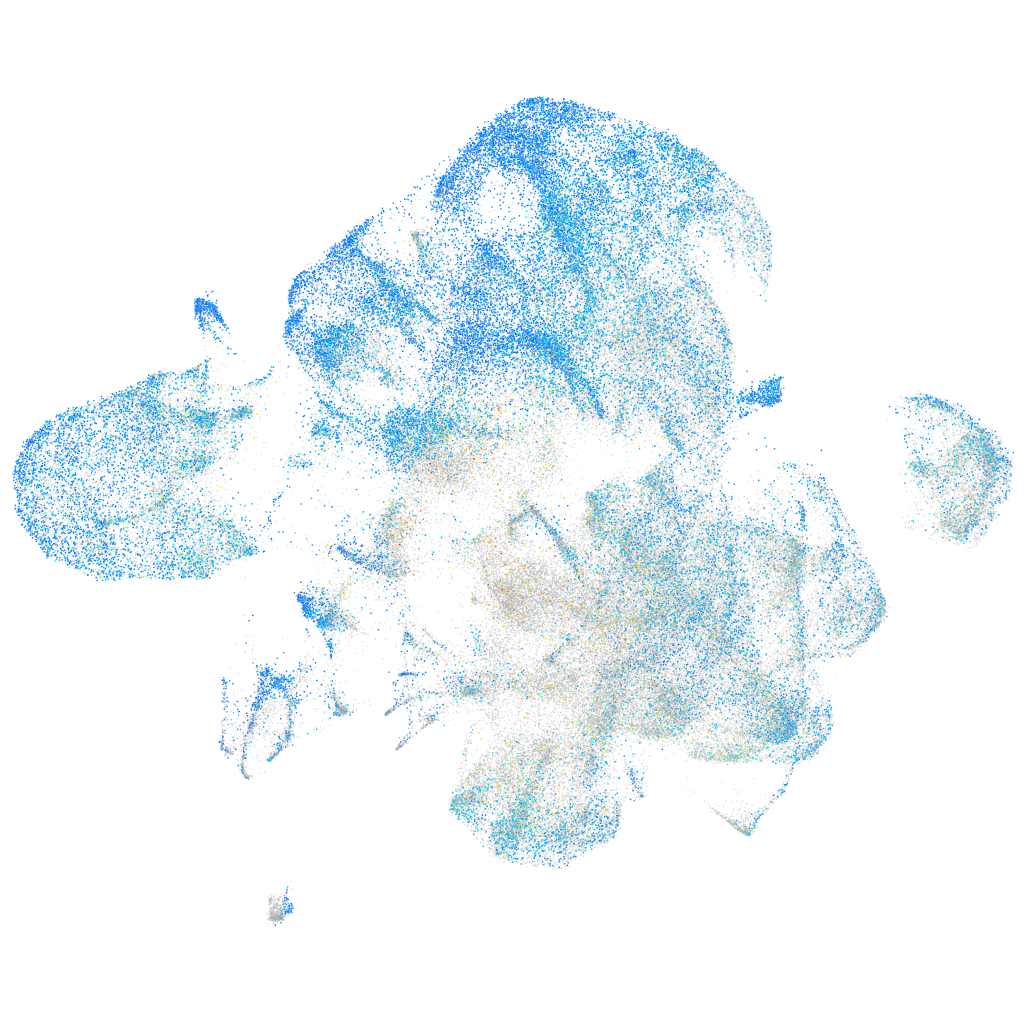
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.229 | rtn1a | -0.159 |
| rrm1 | 0.216 | atp6v0cb | -0.148 |
| hmgb2a | 0.210 | gpm6ab | -0.147 |
| tuba8l4 | 0.204 | gpm6aa | -0.145 |
| anp32b | 0.202 | sncb | -0.143 |
| chaf1a | 0.201 | stmn1b | -0.143 |
| dut | 0.201 | rtn1b | -0.133 |
| ranbp1 | 0.200 | elavl3 | -0.133 |
| mcm7 | 0.198 | elavl4 | -0.133 |
| stmn1a | 0.195 | vamp2 | -0.132 |
| rpa2 | 0.192 | rnasekb | -0.132 |
| rpa3 | 0.191 | gng3 | -0.132 |
| nasp | 0.190 | ywhag2 | -0.126 |
| fen1 | 0.190 | stx1b | -0.124 |
| hmgb2b | 0.189 | ckbb | -0.123 |
| hmga1a | 0.189 | stxbp1a | -0.123 |
| rrm2 | 0.187 | stmn2a | -0.122 |
| ran | 0.186 | si:dkeyp-75h12.5 | -0.121 |
| banf1 | 0.186 | CR383676.1 | -0.119 |
| ccna2 | 0.184 | aplp1 | -0.119 |
| snrpd1 | 0.184 | snap25a | -0.118 |
| dek | 0.184 | atpv0e2 | -0.116 |
| selenoh | 0.183 | pvalb1 | -0.115 |
| id1 | 0.182 | calm1a | -0.114 |
| cbx3a | 0.182 | gap43 | -0.112 |
| nutf2l | 0.180 | atp1a3a | -0.111 |
| seta | 0.178 | zgc:65894 | -0.111 |
| mibp | 0.177 | syt1a | -0.110 |
| slbp | 0.177 | nrxn1a | -0.110 |
| snrpb | 0.176 | sv2a | -0.109 |
| ssrp1a | 0.176 | map1aa | -0.109 |
| lig1 | 0.176 | pvalb2 | -0.108 |
| snrpf | 0.176 | atp6v1e1b | -0.108 |
| zgc:110540 | 0.175 | gapdhs | -0.107 |
| cks1b | 0.174 | olfm1b | -0.106 |