"procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2"
ZFIN 
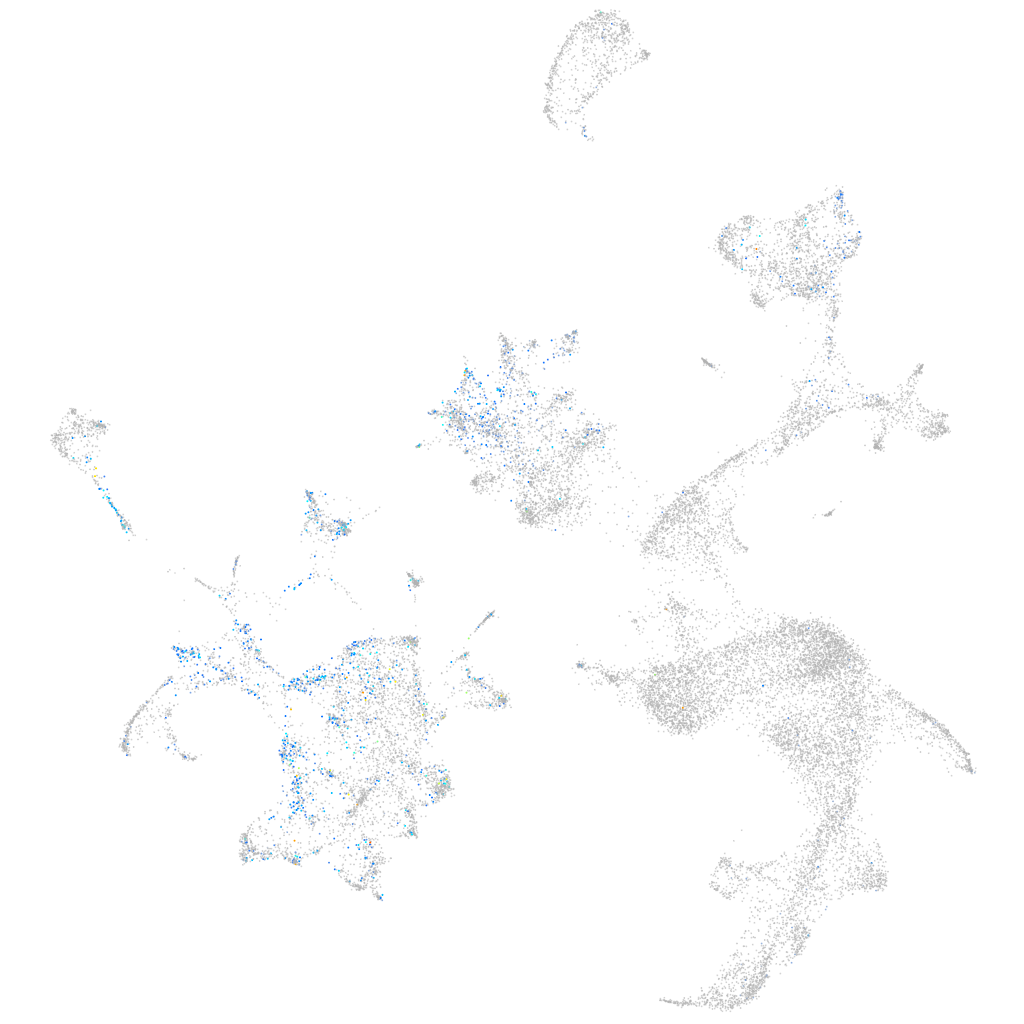
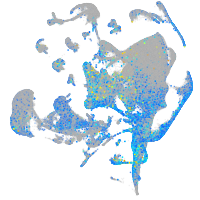
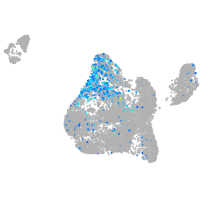
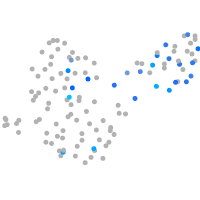
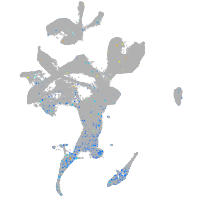
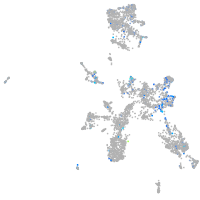
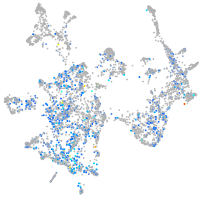
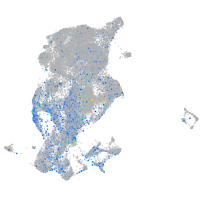
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.205 | prdx2 | -0.144 |
| krt18a.1 | 0.190 | hbae1.1 | -0.139 |
| akap12b | 0.190 | hbae3 | -0.132 |
| p4ha1b | 0.190 | hbbe2 | -0.131 |
| tspan7 | 0.187 | hbbe1.1 | -0.131 |
| etv2 | 0.185 | hbbe1.3 | -0.131 |
| cnn3a | 0.179 | hbae1.3 | -0.129 |
| fkbp14 | 0.179 | cahz | -0.126 |
| fkbp9 | 0.178 | blvrb | -0.126 |
| calua | 0.177 | hemgn | -0.119 |
| ppib | 0.176 | tspo | -0.117 |
| clec14a | 0.176 | slc4a1a | -0.117 |
| dusp5 | 0.175 | alas2 | -0.116 |
| krt8 | 0.172 | hbbe1.2 | -0.116 |
| cd81a | 0.172 | fth1a | -0.110 |
| si:ch211-156j16.1 | 0.171 | epb41b | -0.109 |
| sparc | 0.171 | nt5c2l1 | -0.109 |
| plod1a | 0.170 | zgc:163057 | -0.107 |
| serpinh1b | 0.170 | creg1 | -0.103 |
| colec12 | 0.169 | nmt1b | -0.099 |
| msna | 0.168 | si:ch211-207c6.2 | -0.099 |
| egfl7 | 0.168 | si:ch211-250g4.3 | -0.096 |
| crtap | 0.168 | plac8l1 | -0.091 |
| id1 | 0.165 | gpx1a | -0.088 |
| sept10 | 0.164 | tmod4 | -0.087 |
| marcksl1a | 0.164 | sptb | -0.086 |
| jpt1b | 0.164 | tfr1a | -0.085 |
| kdelr3 | 0.162 | si:ch211-227m13.1 | -0.083 |
| marcksl1b | 0.162 | tmem14ca | -0.083 |
| si:dkey-261h17.1 | 0.159 | hdr | -0.083 |
| fstl1b | 0.158 | hbae5 | -0.079 |
| cdc42ep5 | 0.158 | klf1 | -0.079 |
| fkbp7 | 0.157 | rfesd | -0.077 |
| ramp2 | 0.156 | TCIM (1 of many) | -0.076 |
| cdh5 | 0.156 | ucp3 | -0.076 |