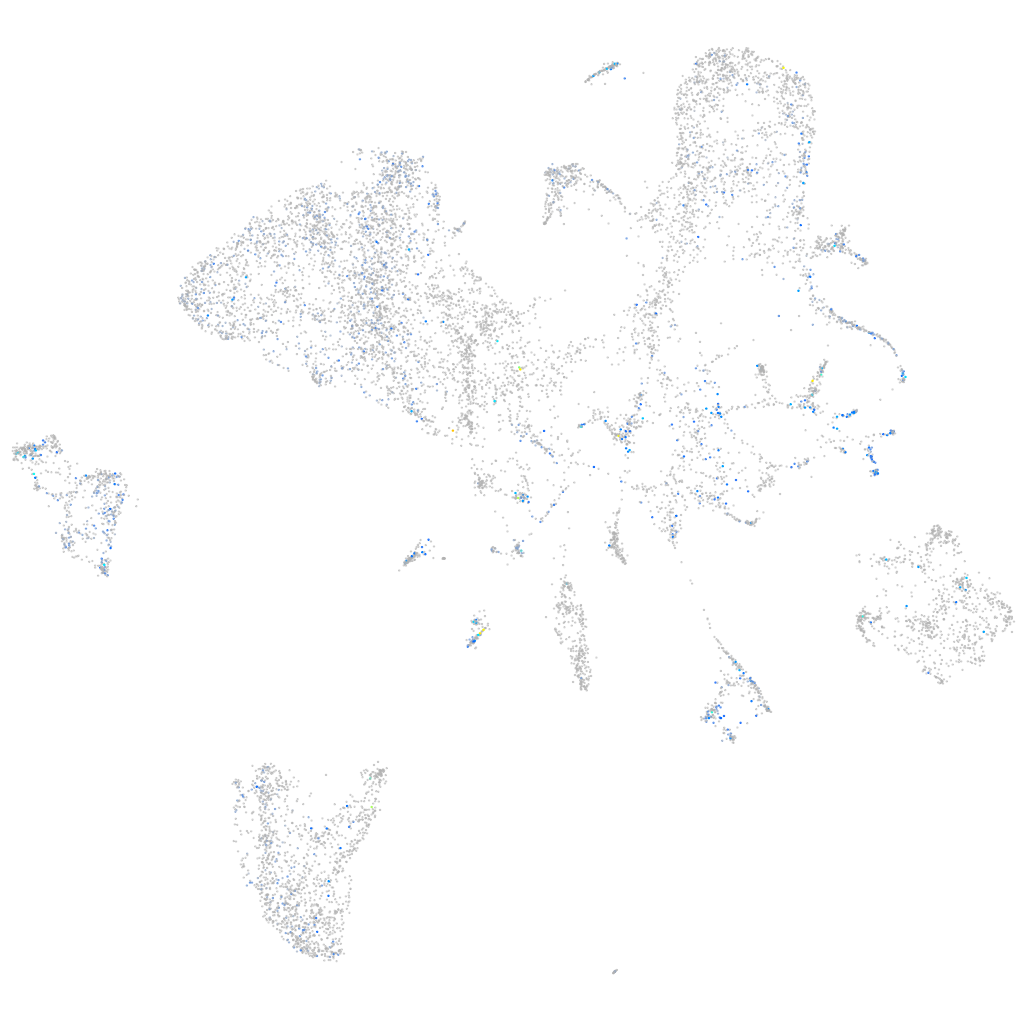
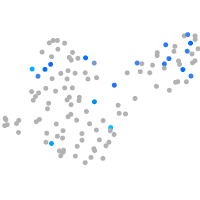
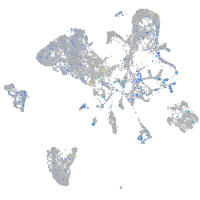
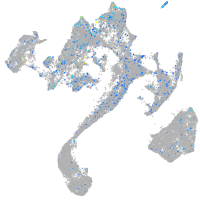
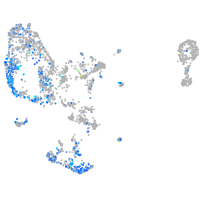
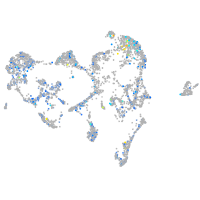
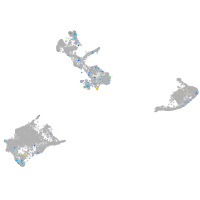
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| areg | 0.136 | gamt | -0.069 |
| pcsk1nl | 0.122 | gapdh | -0.064 |
| cdh16 | 0.121 | apoc2 | -0.061 |
| rims2a | 0.120 | gatm | -0.058 |
| scgn | 0.116 | ahcy | -0.057 |
| gapdhs | 0.116 | fabp1b.1 | -0.056 |
| slc17a7b | 0.116 | afp4 | -0.054 |
| vat1 | 0.115 | rbp2a | -0.053 |
| scg3 | 0.114 | apoc1 | -0.053 |
| spint2 | 0.111 | hspe1 | -0.052 |
| CR383676.1 | 0.109 | apoa4b.1 | -0.052 |
| ptbp3 | 0.108 | apoa1a | -0.051 |
| gcgb | 0.107 | apoa4b.2 | -0.050 |
| cpe | 0.107 | bhmt | -0.049 |
| ier2b | 0.107 | hspb1 | -0.047 |
| LOC101885158 | 0.107 | gstt1a | -0.046 |
| pcsk2 | 0.106 | fbp1b | -0.046 |
| pcsk1 | 0.104 | si:dkey-16p21.8 | -0.046 |
| camk1da | 0.104 | lta4h | -0.046 |
| dkk3b | 0.104 | apoa1b | -0.045 |
| ccni | 0.101 | cdo1 | -0.045 |
| calm1b | 0.101 | scp2a | -0.045 |
| si:dkey-187i8.2 | 0.100 | suclg1 | -0.045 |
| slc30a2 | 0.100 | gpx4a | -0.044 |
| pax6b | 0.100 | zmp:0000000624 | -0.044 |
| rnasekb | 0.099 | fbl | -0.044 |
| kctd12.2 | 0.098 | ing5b | -0.043 |
| pam | 0.098 | aqp12 | -0.043 |
| pnoca | 0.098 | npm1a | -0.043 |
| CR382325.1 | 0.097 | si:ch211-113d11.6 | -0.043 |
| mcl1a | 0.097 | pla2g12b | -0.042 |
| LOC103908954 | 0.097 | si:ch211-201h21.5 | -0.042 |
| zgc:101731 | 0.097 | apoeb | -0.041 |
| pbxip1b | 0.096 | vox | -0.041 |
| tango2 | 0.096 | zgc:112146 | -0.040 |