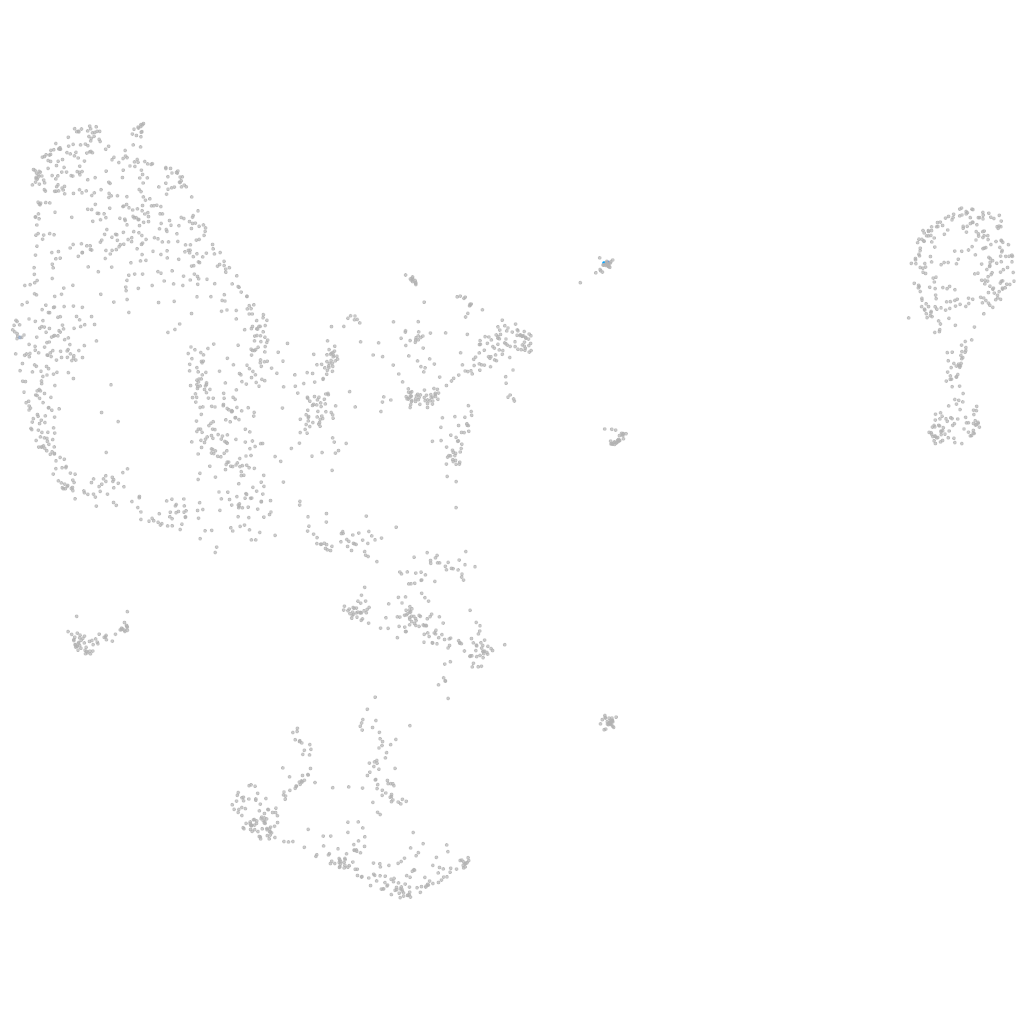
osteocrin
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnmb2 | 0.591 | uqcrfs1 | -0.040 |
| si:ch73-367f21.5 | 0.516 | ndufs4 | -0.039 |
| arhgap44 | 0.473 | hspe1 | -0.037 |
| irf4b | 0.436 | ddx39ab | -0.037 |
| bhlha15 | 0.408 | bud31 | -0.035 |
| zgc:152753 | 0.400 | rps18 | -0.035 |
| si:ch211-154e10.1 | 0.396 | mrpl12 | -0.034 |
| XLOC-005448 | 0.396 | ndufb9 | -0.034 |
| si:dkey-103e21.5 | 0.363 | snrpd1 | -0.033 |
| arfip2b | 0.360 | ndufa7 | -0.033 |
| stat4 | 0.350 | aldob | -0.032 |
| XLOC-026741 | 0.335 | ppp2cb | -0.030 |
| LOC108179070 | 0.334 | tuba8l | -0.030 |
| hoxa1a | 0.325 | eif5a2 | -0.030 |
| swsap1 | 0.318 | pa2g4b | -0.030 |
| HTRA2 (1 of many).4 | 0.310 | eif4g1a | -0.030 |
| arhgef10 | 0.300 | hnrnpa1b | -0.030 |
| lin7a | 0.293 | cdaa | -0.030 |
| camsap2b | 0.290 | emc10 | -0.029 |
| hcst | 0.289 | ehd1b | -0.029 |
| CT030153.1 | 0.288 | c1qbp | -0.029 |
| stc1l | 0.284 | cotl1 | -0.029 |
| rhobtb2b | 0.282 | nop56 | -0.029 |
| rem1 | 0.280 | timm9 | -0.029 |
| anxa5a | 0.279 | zgc:101040 | -0.029 |
| tns2a | 0.277 | ndufa6 | -0.028 |
| mef2ca | 0.275 | kars | -0.028 |
| sema3bl | 0.269 | txn2 | -0.028 |
| pde4ca | 0.266 | cox4i1 | -0.028 |
| LOC101883751 | 0.266 | tomm5 | -0.028 |
| LOC570115 | 0.265 | ddt | -0.027 |
| zgc:158464 | 0.263 | si:dkey-56m19.5 | -0.027 |
| npdc1a | 0.260 | clptm1 | -0.027 |
| CABZ01102518.1 | 0.259 | mrps11 | -0.027 |
| tnfb | 0.253 | ndufab1a | -0.027 |