opiate receptor-like 1
ZFIN 
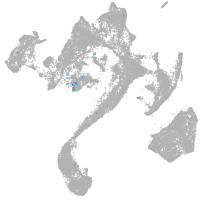
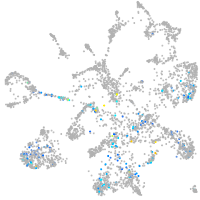
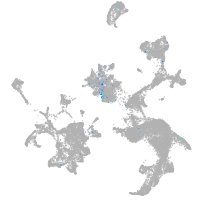
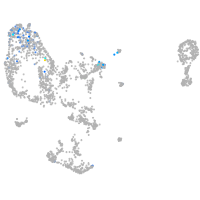
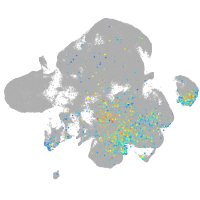
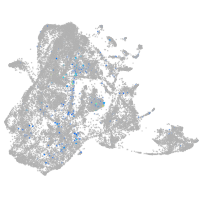
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg2b | 0.195 | cx43.4 | -0.074 |
| ywhag2 | 0.185 | hspb1 | -0.073 |
| sncb | 0.182 | akap12b | -0.068 |
| snap25a | 0.181 | stm | -0.067 |
| zgc:65894 | 0.176 | anp32e | -0.065 |
| stmn2a | 0.175 | msna | -0.065 |
| sv2a | 0.175 | si:ch211-152c2.3 | -0.065 |
| slc6a1a | 0.172 | pou5f3 | -0.064 |
| stx1b | 0.168 | dkc1 | -0.063 |
| elavl4 | 0.166 | id1 | -0.063 |
| atp6v0cb | 0.165 | mki67 | -0.063 |
| stxbp1a | 0.163 | nop58 | -0.063 |
| rtn1b | 0.161 | apoeb | -0.062 |
| syngr3a | 0.160 | ccng1 | -0.062 |
| sypa | 0.159 | hnrnpa1b | -0.062 |
| cplx2 | 0.157 | npm1a | -0.062 |
| gap43 | 0.157 | banf1 | -0.061 |
| map1aa | 0.157 | fbl | -0.061 |
| aplp1 | 0.156 | s100a1 | -0.061 |
| vamp2 | 0.156 | arf1 | -0.060 |
| rab6bb | 0.154 | polr3gla | -0.060 |
| camk2n1a | 0.153 | chaf1a | -0.059 |
| eno2 | 0.153 | lig1 | -0.059 |
| tmsb2 | 0.152 | zgc:56699 | -0.059 |
| vsnl1b | 0.152 | ucp2 | -0.058 |
| syn2a | 0.151 | ccnb1 | -0.057 |
| basp1 | 0.150 | si:dkey-66i24.9 | -0.057 |
| cdk5r2a | 0.148 | tpx2 | -0.057 |
| nrxn1a | 0.145 | zgc:110425 | -0.057 |
| BX005003.1 | 0.143 | asb11 | -0.056 |
| nova1 | 0.143 | bzw1b | -0.056 |
| atp2b3b | 0.142 | eif5a2 | -0.056 |
| sprn | 0.142 | nop56 | -0.056 |
| ywhag1 | 0.141 | apoc1 | -0.055 |
| gng3 | 0.140 | ccna2 | -0.055 |