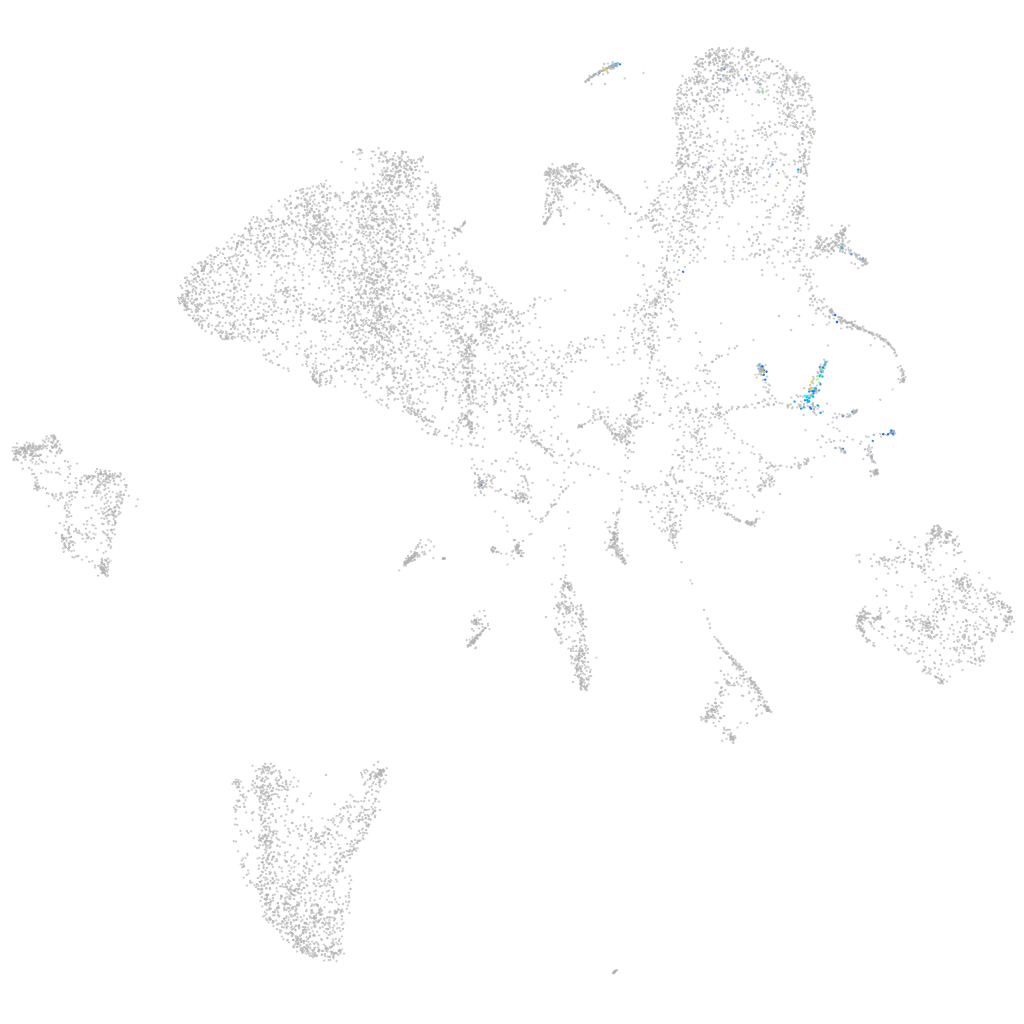
"opioid receptor, delta 1b"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| arxa | 0.379 | ahcy | -0.098 |
| scg3 | 0.366 | fabp3 | -0.098 |
| si:ch73-160i9.2 | 0.345 | rps6 | -0.084 |
| scgn | 0.344 | rpl6 | -0.083 |
| rasd4 | 0.313 | gamt | -0.080 |
| ccka | 0.307 | bhmt | -0.080 |
| egr4 | 0.304 | eif4ebp1 | -0.078 |
| c2cd4a | 0.300 | hdlbpa | -0.078 |
| insl5a | 0.299 | gatm | -0.074 |
| fev | 0.295 | gapdh | -0.074 |
| slc35g2a | 0.295 | ppiaa | -0.072 |
| plcxd3 | 0.290 | mat1a | -0.072 |
| LOC101885494 | 0.288 | si:dkey-16p21.8 | -0.070 |
| insm1b | 0.283 | apoc1 | -0.069 |
| calca | 0.283 | aqp12 | -0.069 |
| pcsk2 | 0.273 | rps20 | -0.069 |
| rem2 | 0.273 | eef2b | -0.068 |
| rprmb | 0.273 | sec61a1 | -0.067 |
| kcnh6a | 0.271 | eif5a | -0.067 |
| neurod1 | 0.269 | aldh6a1 | -0.066 |
| calm1b | 0.265 | cx32.3 | -0.066 |
| vat1 | 0.263 | adka | -0.065 |
| tango2 | 0.261 | agxta | -0.065 |
| tspan7b | 0.260 | agxtb | -0.065 |
| mctp2b | 0.260 | apoa4b.1 | -0.064 |
| hepacam2 | 0.259 | tktb | -0.064 |
| npy2rl | 0.259 | apoa1b | -0.063 |
| galn | 0.255 | gstt1a | -0.063 |
| anxa5a | 0.253 | wu:fj16a03 | -0.063 |
| capsla | 0.253 | shmt1 | -0.063 |
| elovl1a | 0.251 | gnmt | -0.063 |
| pcsk1nl | 0.250 | afp4 | -0.062 |
| dzip1l | 0.250 | aldh7a1 | -0.061 |
| cplx2 | 0.249 | tfa | -0.060 |
| dll4 | 0.248 | serpina1 | -0.060 |