olfactomedin 3b
ZFIN 
Other cell groups


all cells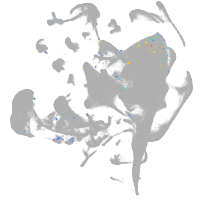 |
blastomeres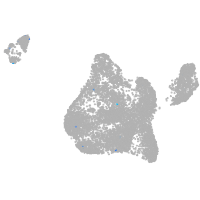 |
PGCs |
endoderm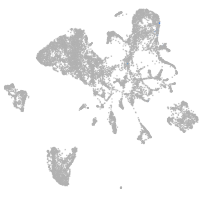 |
axial mesoderm |
muscle 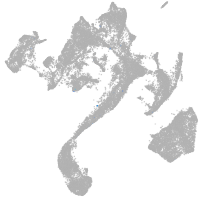 |
mural cells / non-skeletal muscle 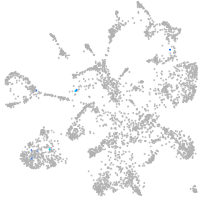 |
mesenchyme 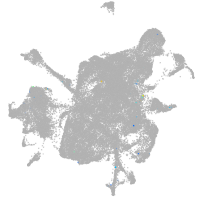 |
hematopoietic / vasculature 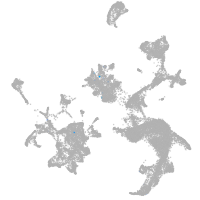 |
pronephros  |
neural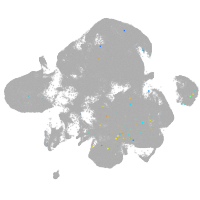 |
spinal cord / glia |
eye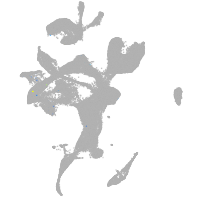 |
taste / olfactory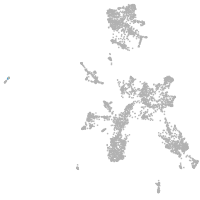 |
otic / lateral line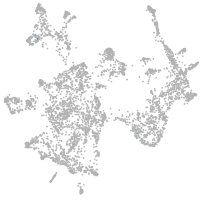 |
epidermis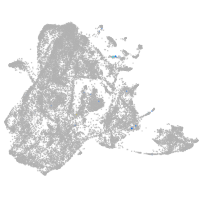 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886621 | 0.051 | cx43.4 | -0.012 |
| trpv6 | 0.050 | stm | -0.012 |
| FP016234.1 | 0.044 | aldob | -0.011 |
| ywhag2 | 0.044 | apoc1 | -0.011 |
| atp6v0cb | 0.038 | hspb1 | -0.011 |
| atpv0e2 | 0.038 | lig1 | -0.011 |
| map1aa | 0.038 | si:ch211-152c2.3 | -0.011 |
| nptna | 0.038 | apoeb | -0.010 |
| rprml | 0.038 | cast | -0.010 |
| scg2b | 0.038 | dkc1 | -0.010 |
| sncb | 0.038 | dnmt3bb.2 | -0.010 |
| stmn2a | 0.038 | fbl | -0.010 |
| syn2a | 0.038 | galnt7 | -0.010 |
| ywhag1 | 0.038 | gsnb | -0.010 |
| zgc:65894 | 0.038 | si:dkey-66i24.9 | -0.010 |
| calm3a | 0.038 | sox19a | -0.010 |
| camk2n1a | 0.037 | aep1 | -0.009 |
| cdk5r2a | 0.037 | akap12b | -0.009 |
| snap25a | 0.037 | arf1 | -0.009 |
| vamp2 | 0.037 | banf1 | -0.009 |
| atp6v1b2 | 0.036 | BX927258.1 | -0.009 |
| fxyd6 | 0.036 | capn9 | -0.009 |
| si:ch73-119p20.1 | 0.036 | ccnd1 | -0.009 |
| LO017848.1 | 0.035 | chaf1a | -0.009 |
| npas4a | 0.035 | cyp2y3 | -0.009 |
| atp2b3b | 0.034 | cyt1l | -0.009 |
| calm1b | 0.034 | gnl3 | -0.009 |
| eno2 | 0.034 | hnrnpa1b | -0.009 |
| olfm2a | 0.034 | hnrnpm | -0.009 |
| rnasekb | 0.034 | id1 | -0.009 |
| stmn4 | 0.034 | lye | -0.009 |
| vsnl1b | 0.034 | mcm3 | -0.009 |
| calm1a | 0.034 | mcm6 | -0.009 |
| atp6v1e1b | 0.033 | npm1a | -0.009 |
| egr4 | 0.033 | polr3gla | -0.009 |




