oculocutaneous albinism II
ZFIN 
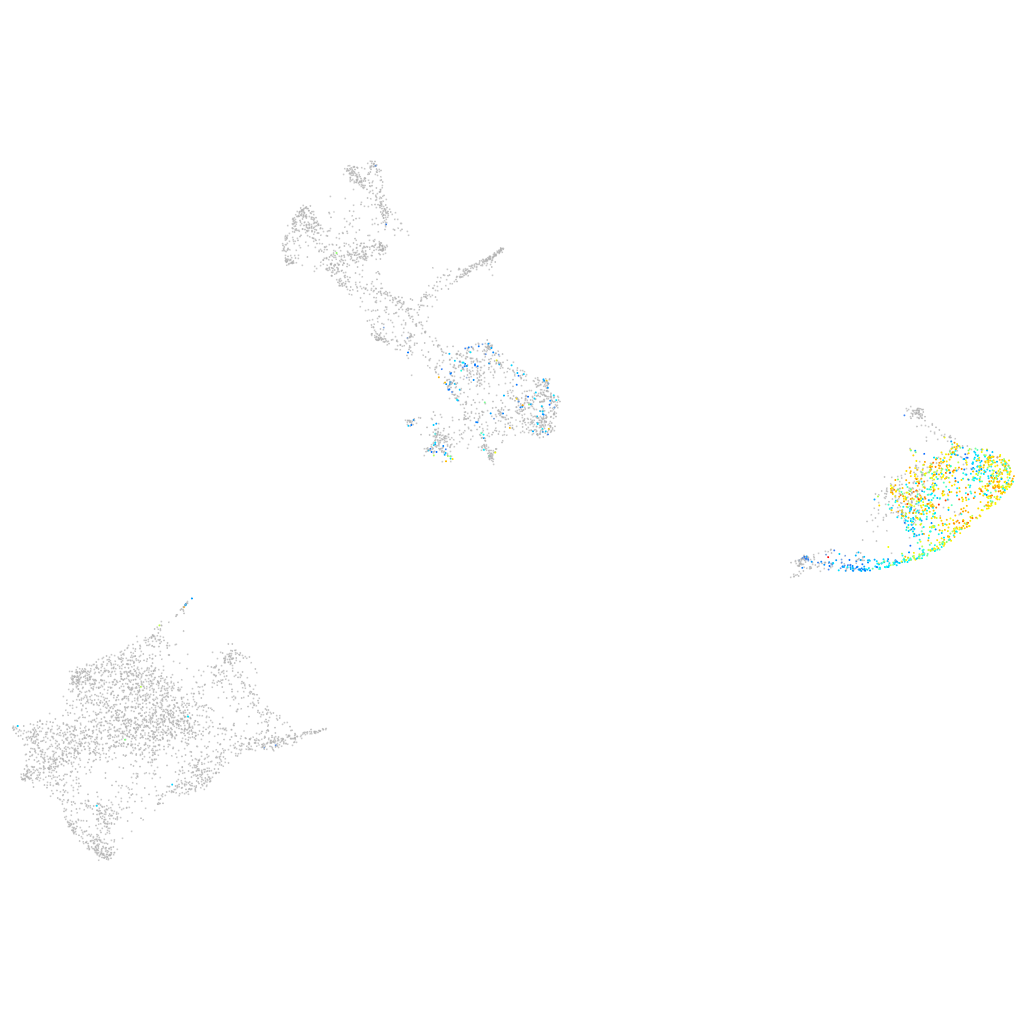
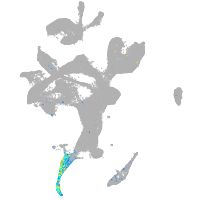
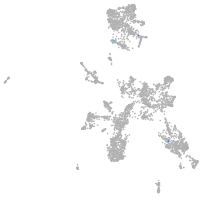
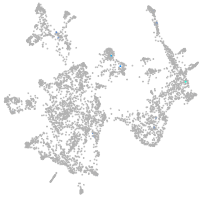
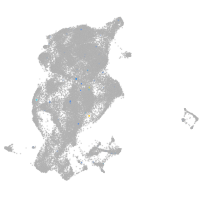
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1b | 0.845 | paics | -0.467 |
| pmela | 0.844 | CABZ01021592.1 | -0.444 |
| tyrp1a | 0.837 | uraha | -0.387 |
| dct | 0.818 | mdh1aa | -0.351 |
| zgc:91968 | 0.786 | si:dkey-251i10.2 | -0.348 |
| mtbl | 0.719 | impdh1b | -0.330 |
| kita | 0.705 | tmem130 | -0.326 |
| slc24a5 | 0.700 | slc2a15a | -0.318 |
| prkar1b | 0.698 | glulb | -0.315 |
| tyr | 0.666 | atic | -0.310 |
| slc45a2 | 0.647 | phyhd1 | -0.293 |
| si:ch73-389b16.1 | 0.642 | aox5 | -0.284 |
| slc39a10 | 0.635 | si:ch211-251b21.1 | -0.276 |
| slc22a2 | 0.623 | prps1a | -0.271 |
| aadac | 0.618 | cyb5a | -0.248 |
| kcnj13 | 0.604 | bscl2l | -0.248 |
| anxa1a | 0.604 | ppat | -0.241 |
| gstt1a | 0.593 | pax7b | -0.240 |
| slc29a3 | 0.567 | krt18b | -0.234 |
| lamp1a | 0.531 | pax7a | -0.232 |
| SPAG9 | 0.518 | gmps | -0.232 |
| slc7a5 | 0.515 | shmt1 | -0.231 |
| hsd20b2 | 0.513 | sprb | -0.226 |
| mlpha | 0.505 | bco1 | -0.219 |
| tspan36 | 0.499 | cx30.3 | -0.217 |
| slc37a2 | 0.498 | aldob | -0.214 |
| zdhhc2 | 0.495 | cax1 | -0.214 |
| tspan10 | 0.494 | TMEM19 | -0.214 |
| slc24a4a | 0.494 | sult1st1 | -0.213 |
| slc3a2a | 0.492 | prdx1 | -0.207 |
| CDK18 | 0.490 | si:ch211-194k22.8 | -0.205 |
| s100a10b | 0.483 | defbl1 | -0.203 |
| atp6v0ca | 0.482 | apoda.1 | -0.201 |
| slc30a1b | 0.476 | mibp | -0.201 |
| spock3 | 0.474 | prdx5 | -0.198 |