nucleoporin 188
ZFIN 
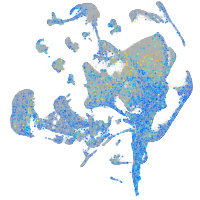
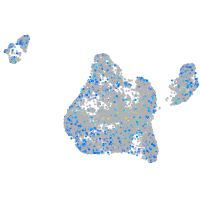
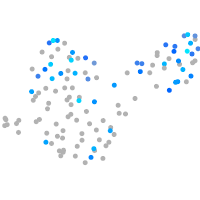
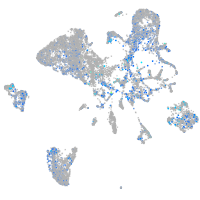
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.128 | pvalb1 | -0.054 |
| anp32b | 0.113 | pvalb2 | -0.052 |
| nop58 | 0.111 | actc1b | -0.049 |
| tpx2 | 0.111 | ckbb | -0.048 |
| fbl | 0.109 | rtn1a | -0.045 |
| ranbp1 | 0.109 | atp6v0cb | -0.044 |
| banf1 | 0.108 | gpm6ab | -0.042 |
| nop56 | 0.108 | rnasekb | -0.042 |
| snrpb | 0.108 | stmn1b | -0.042 |
| ccnb1 | 0.107 | gng3 | -0.041 |
| dkc1 | 0.107 | sncb | -0.040 |
| snu13b | 0.107 | calm1a | -0.039 |
| chaf1a | 0.106 | atp6v1e1b | -0.037 |
| mad2l1 | 0.106 | ccni | -0.037 |
| pcna | 0.106 | mylpfa | -0.037 |
| seta | 0.105 | rbp4 | -0.037 |
| ccna2 | 0.104 | si:dkeyp-75h12.5 | -0.037 |
| ncl | 0.104 | ak1 | -0.036 |
| eif5a2 | 0.103 | atpv0e2 | -0.036 |
| lig1 | 0.102 | hbbe1.3 | -0.036 |
| npm1a | 0.102 | rtn1b | -0.036 |
| rrm1 | 0.102 | vamp2 | -0.036 |
| snrpd1 | 0.102 | hbae3 | -0.035 |
| top2a | 0.101 | tpi1b | -0.035 |
| ccnd1 | 0.100 | gabarapl2 | -0.034 |
| hnrnpa1b | 0.100 | gapdhs | -0.034 |
| plk1 | 0.100 | gpm6aa | -0.034 |
| ppm1g | 0.100 | mylz3 | -0.034 |
| cbx3a | 0.099 | ywhag2 | -0.034 |
| dek | 0.099 | elavl3 | -0.033 |
| mcm7 | 0.099 | elavl4 | -0.033 |
| smc2 | 0.099 | gabarapa | -0.033 |
| ube2c | 0.099 | hbbe1.1 | -0.033 |
| aurkb | 0.097 | olfm1b | -0.033 |
| cdca8 | 0.097 | vdac3 | -0.033 |