nucleoporin 153
ZFIN 
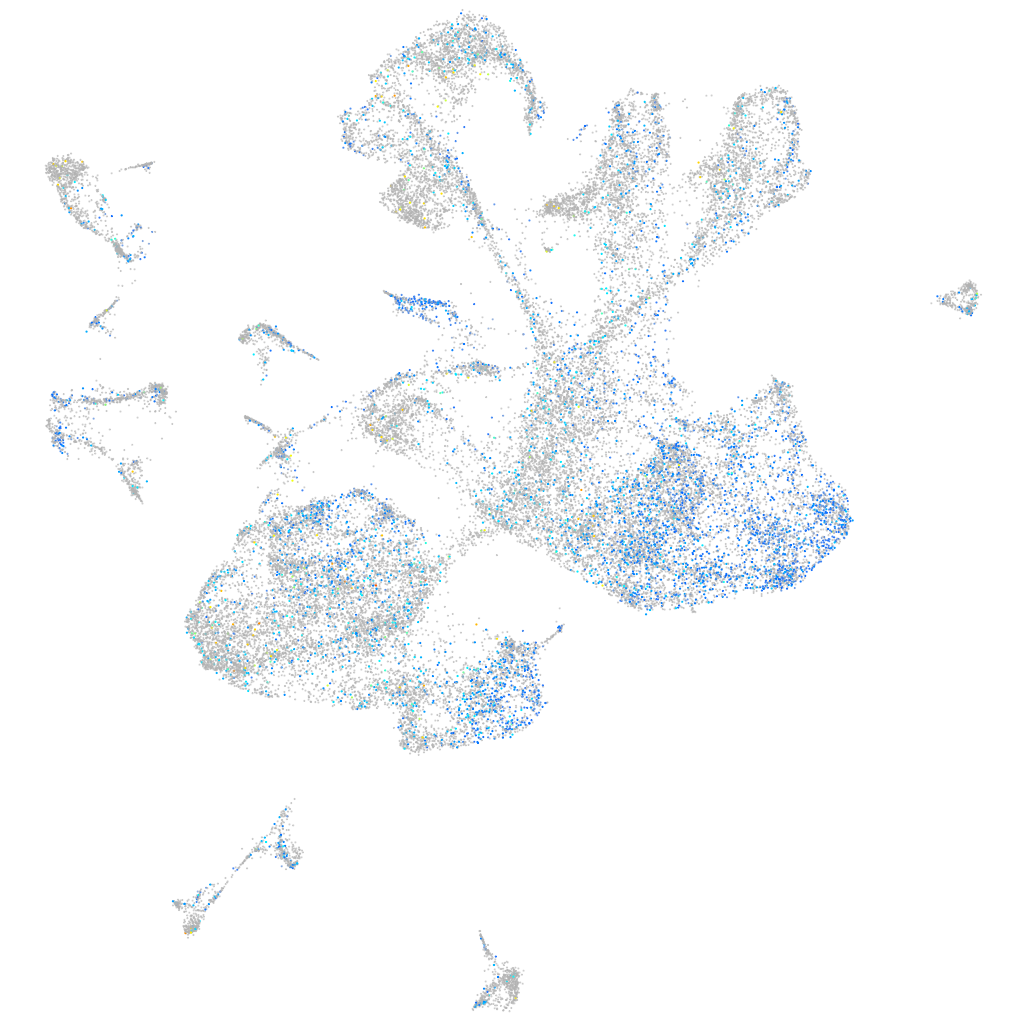
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.150 | ppdpfb | -0.081 |
| rrm1 | 0.147 | pvalb2 | -0.067 |
| chaf1a | 0.146 | CR383676.1 | -0.066 |
| dut | 0.145 | ckbb | -0.066 |
| rpa2 | 0.139 | actc1b | -0.065 |
| rrm2 | 0.138 | gpm6ab | -0.064 |
| lig1 | 0.138 | rtn1a | -0.063 |
| mibp | 0.137 | pvalb1 | -0.062 |
| zgc:110216 | 0.133 | si:ch211-260e23.9 | -0.062 |
| zgc:110540 | 0.133 | stmn1b | -0.062 |
| dek | 0.132 | si:dkeyp-75h12.5 | -0.061 |
| fen1 | 0.132 | elavl4 | -0.060 |
| hmgb2a | 0.132 | hbae3 | -0.059 |
| banf1 | 0.131 | rnasekb | -0.057 |
| nasp | 0.130 | calm1a | -0.057 |
| CABZ01005379.1 | 0.130 | atp6v0cb | -0.057 |
| ranbp1 | 0.130 | cadm4 | -0.056 |
| anp32b | 0.129 | gpm6aa | -0.056 |
| ssrp1a | 0.127 | elavl3 | -0.055 |
| cks1b | 0.126 | myt1b | -0.055 |
| baz1b | 0.126 | snap25a | -0.055 |
| nop58 | 0.125 | gng3 | -0.055 |
| slbp | 0.125 | tp53inp1 | -0.054 |
| tuba8l4 | 0.124 | sncb | -0.054 |
| mcm7 | 0.123 | gapdhs | -0.052 |
| lbr | 0.123 | vamp2 | -0.050 |
| snrpd1 | 0.122 | cotl1 | -0.049 |
| sumo3b | 0.122 | stx1b | -0.049 |
| stmn1a | 0.122 | hbbe1.1 | -0.049 |
| rnaseh2a | 0.122 | acbd7 | -0.048 |
| nutf2l | 0.121 | dpysl3 | -0.048 |
| npm1a | 0.120 | stxbp1a | -0.048 |
| ccna2 | 0.119 | hbbe1.3 | -0.048 |
| ppm1g | 0.119 | gabarapl2 | -0.048 |
| fbxo5 | 0.119 | mylpfa | -0.047 |