"5'-nucleotidase, cytosolic IIb"
ZFIN 
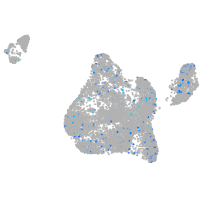
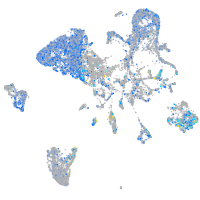
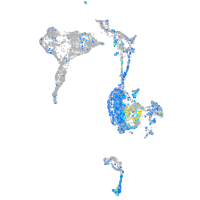
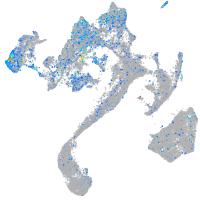
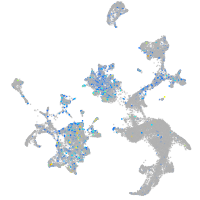
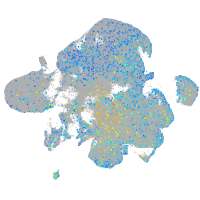
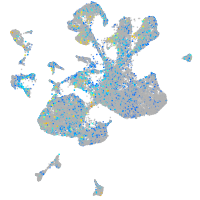
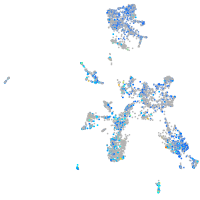
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.259 | paics | -0.141 |
| kita | 0.242 | atic | -0.120 |
| CR847566.1 | 0.215 | impdh1b | -0.112 |
| oca2 | 0.208 | uraha | -0.099 |
| slc24a5 | 0.207 | CABZ01021592.1 | -0.098 |
| tyrp1a | 0.207 | mdh1aa | -0.094 |
| pmela | 0.201 | prps1a | -0.091 |
| si:ch211-195b13.1 | 0.201 | glulb | -0.087 |
| slc22a2 | 0.200 | si:dkey-151g10.6 | -0.079 |
| tyrp1b | 0.199 | defbl1 | -0.076 |
| slc24a4a | 0.196 | si:dkey-251i10.2 | -0.076 |
| dct | 0.194 | apoda.1 | -0.074 |
| slc39a10 | 0.193 | shmt1 | -0.072 |
| lamp1a | 0.186 | phyhd1 | -0.072 |
| slc30a1b | 0.185 | aqp1a.1 | -0.071 |
| ocstamp | 0.185 | si:ch211-256m1.8 | -0.070 |
| aadac | 0.185 | ppat | -0.069 |
| slc7a5 | 0.182 | tmem130 | -0.068 |
| zdhhc2 | 0.179 | gpnmb | -0.068 |
| tyr | 0.177 | si:dkey-197i20.6 | -0.067 |
| ccng2 | 0.176 | gmps | -0.066 |
| mtbl | 0.176 | tfec | -0.066 |
| si:ch73-389b16.1 | 0.174 | lypc | -0.066 |
| slc43a2a | 0.174 | unm-sa821 | -0.065 |
| sik1 | 0.172 | rbp4l | -0.065 |
| slc29a3 | 0.171 | rps5 | -0.064 |
| slc7a8a | 0.169 | cyb5a | -0.063 |
| prkar1b | 0.169 | gart | -0.063 |
| CR383676.1 | 0.169 | guk1a | -0.063 |
| slc30a8 | 0.167 | akap12a | -0.061 |
| AL935199.1 | 0.167 | rps7 | -0.061 |
| kcnj13 | 0.164 | psat1 | -0.061 |
| rap1gap | 0.164 | s100v2 | -0.060 |
| mcf2l2 | 0.164 | ponzr1 | -0.059 |
| zgc:91968 | 0.164 | krt18b | -0.059 |