NSL1 component of MIS12 kinetochore complex
ZFIN 
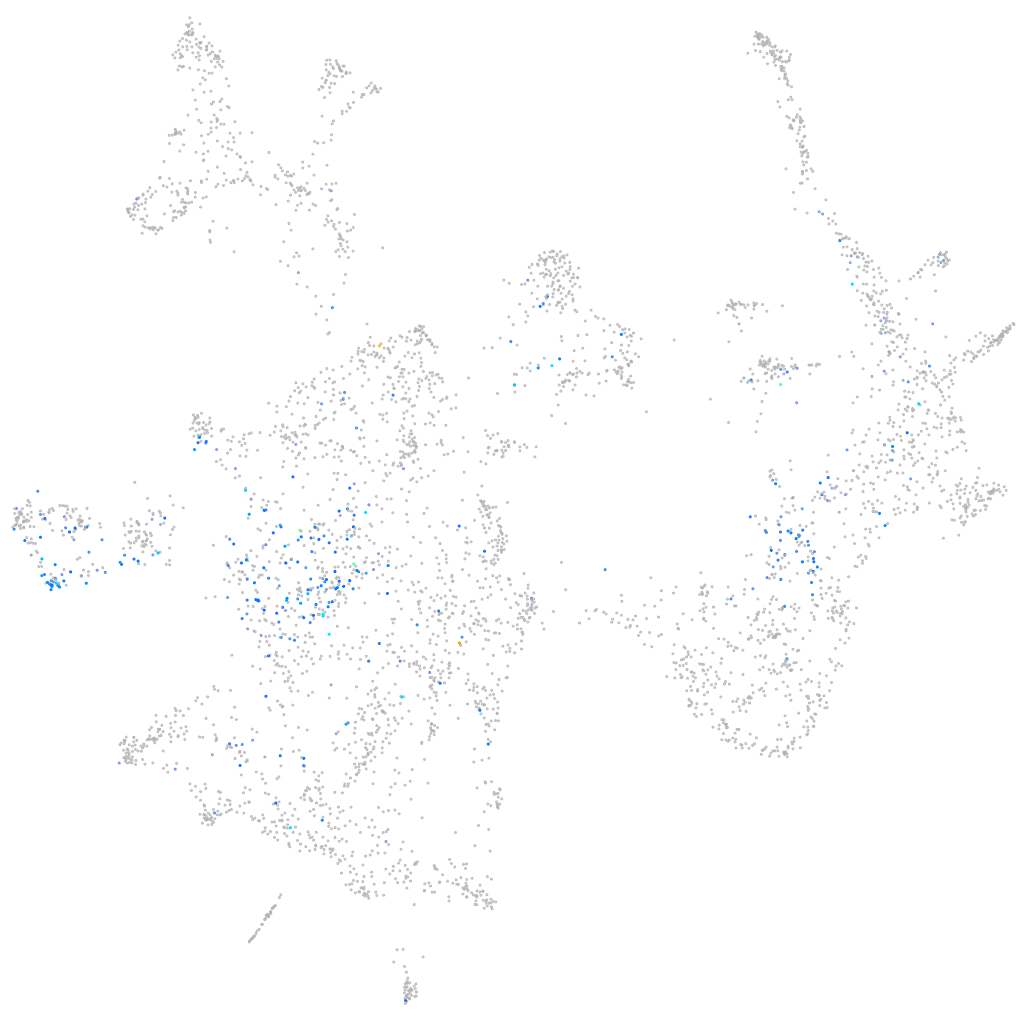
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mad2l1 | 0.373 | COX3 | -0.109 |
| cdk1 | 0.363 | tob1b | -0.101 |
| cks1b | 0.359 | pvalb2 | -0.100 |
| ccna2 | 0.352 | CR383676.1 | -0.098 |
| cdca5 | 0.347 | tmem59 | -0.098 |
| aurkb | 0.337 | gabarapl2 | -0.095 |
| mki67 | 0.334 | atp2b1a | -0.094 |
| rrm1 | 0.332 | pvalb1 | -0.092 |
| plk1 | 0.330 | atp6ap2 | -0.091 |
| nuf2 | 0.324 | eno1a | -0.091 |
| zgc:110216 | 0.322 | mt-co2 | -0.090 |
| dut | 0.321 | b2ml | -0.090 |
| ccnb1 | 0.315 | rnasekb | -0.090 |
| tpx2 | 0.314 | kif1aa | -0.090 |
| ncapd2 | 0.310 | nptna | -0.089 |
| top2a | 0.307 | otofb | -0.088 |
| ncapg | 0.299 | calml4a | -0.087 |
| si:dkey-6i22.5 | 0.299 | dnajc5b | -0.087 |
| zgc:165555.3 | 0.297 | pvalb8 | -0.087 |
| dek | 0.297 | pkig | -0.086 |
| tubb2b | 0.296 | gpx2 | -0.086 |
| mibp | 0.293 | CR925719.1 | -0.086 |
| dhfr | 0.292 | cd164l2 | -0.086 |
| sgo1 | 0.291 | calm1a | -0.086 |
| nusap1 | 0.290 | atp1a3b | -0.085 |
| dlgap5 | 0.290 | nupr1a | -0.085 |
| stmn1a | 0.290 | pcsk5a | -0.083 |
| si:dkey-261m9.17 | 0.288 | zgc:158463 | -0.083 |
| kifc1 | 0.287 | rplp2 | -0.082 |
| cenpn | 0.285 | map1lc3a | -0.082 |
| rpa2 | 0.284 | gapdhs | -0.082 |
| smc2 | 0.284 | aldocb | -0.082 |
| kif22 | 0.283 | atp6v1e1b | -0.082 |
| ncaph | 0.282 | s100t | -0.081 |
| fbxo5 | 0.282 | twf2b | -0.081 |