"nuclear receptor subfamily 2, group F, member 6b"
ZFIN 
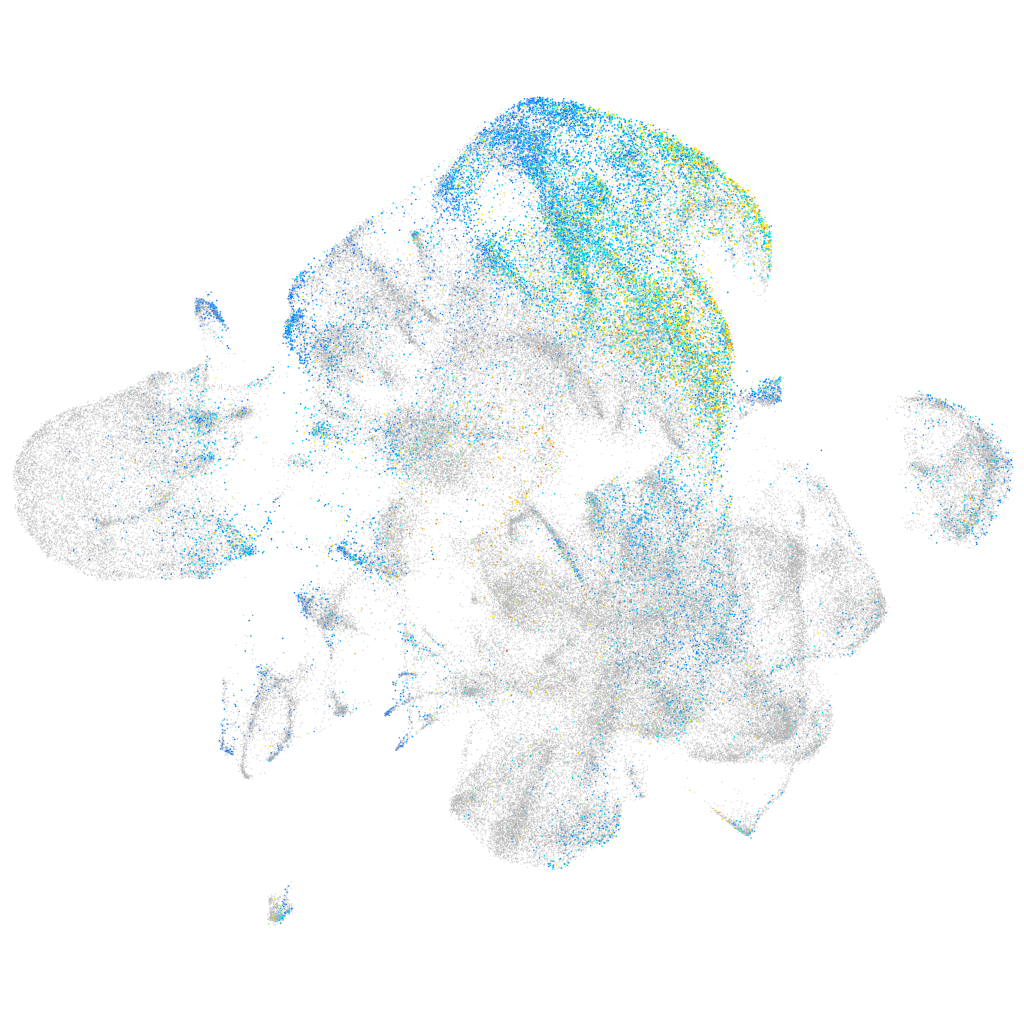
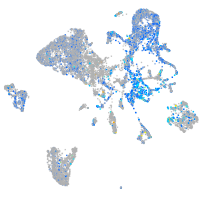
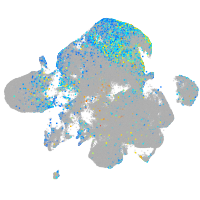
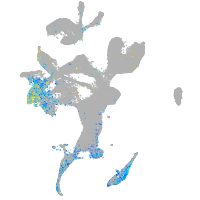
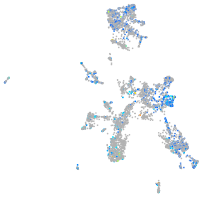
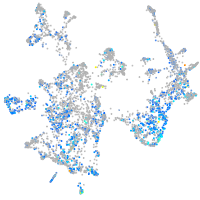
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fstl1a | 0.387 | ckbb | -0.199 |
| si:ch211-57n23.4 | 0.351 | gpm6ab | -0.182 |
| gfap | 0.347 | stmn1b | -0.182 |
| mir219-3 | 0.331 | gng3 | -0.171 |
| XLOC-003689 | 0.327 | sncb | -0.164 |
| CABZ01075068.1 | 0.326 | elavl4 | -0.163 |
| sb:cb81 | 0.316 | rtn1b | -0.161 |
| si:ch73-21g5.7 | 0.314 | atp6v0cb | -0.152 |
| cldn5a | 0.313 | fez1 | -0.146 |
| sertad2a | 0.311 | stx1b | -0.141 |
| LOC798783 | 0.304 | stmn2a | -0.139 |
| lrrn1 | 0.302 | ywhag2 | -0.138 |
| XLOC-003690 | 0.287 | gapdhs | -0.137 |
| notch3 | 0.283 | vamp2 | -0.135 |
| lfng | 0.282 | sox4a | -0.132 |
| sox3 | 0.281 | zgc:65894 | -0.131 |
| dld | 0.277 | snap25a | -0.130 |
| her12 | 0.273 | atp6v1e1b | -0.129 |
| plp1a | 0.272 | olfm1b | -0.127 |
| CU633775.1 | 0.269 | tmsb2 | -0.127 |
| nrarpb | 0.259 | myt1la | -0.126 |
| btg2 | 0.259 | aplp1 | -0.126 |
| gadd45gb.1 | 0.254 | rnasekb | -0.126 |
| tspan7 | 0.253 | cbx1b | -0.125 |
| cep131 | 0.246 | tmem59l | -0.124 |
| angptl4 | 0.244 | csdc2a | -0.123 |
| dla | 0.243 | stxbp1a | -0.123 |
| gpm6bb | 0.243 | map1aa | -0.123 |
| slc1a3a | 0.239 | pvalb1 | -0.123 |
| nes | 0.239 | gap43 | -0.121 |
| si:ch211-286o17.1 | 0.237 | cdk5r1b | -0.121 |
| si:dkey-42i9.4 | 0.235 | si:dkeyp-75h12.5 | -0.118 |
| id1 | 0.234 | rbfox1 | -0.118 |
| prtgb | 0.231 | atpv0e2 | -0.118 |
| her15.1 | 0.228 | calm2a | -0.118 |