nicotinamide nucleotide transhydrogenase
ZFIN 
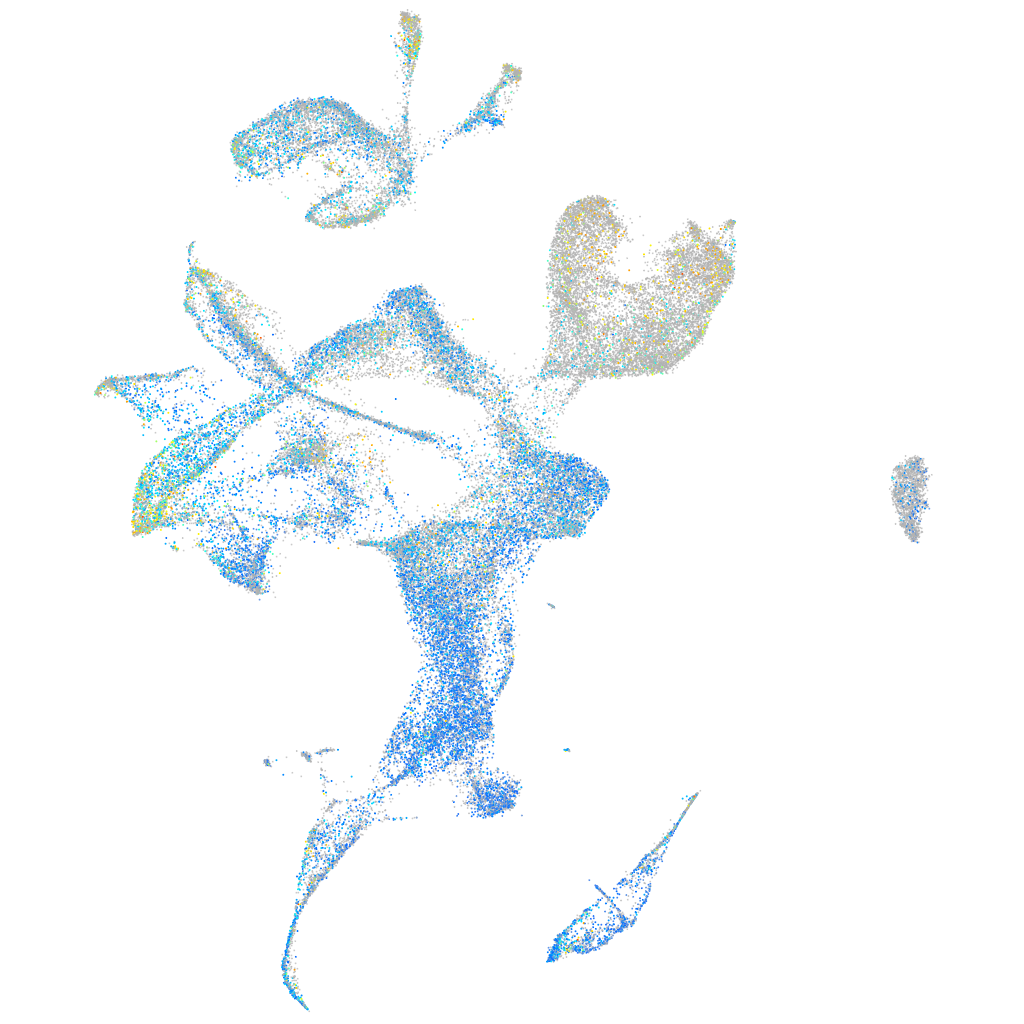
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gnb3b | 0.167 | vsx1 | -0.103 |
| gnat2 | 0.166 | gnb3a | -0.100 |
| pde6c | 0.166 | marcksl1b | -0.096 |
| prph2a | 0.165 | ptmab | -0.091 |
| slc25a3a | 0.163 | mdkb | -0.088 |
| grk1b | 0.159 | calb2b | -0.087 |
| guk1b | 0.159 | h3f3d | -0.084 |
| prph2b | 0.159 | pcp4l1 | -0.081 |
| grk7a | 0.158 | nrn1lb | -0.081 |
| pdcb | 0.158 | gng13b | -0.080 |
| zgc:73359 | 0.155 | si:ch1073-83n3.2 | -0.080 |
| si:dkey-17e16.15 | 0.153 | bcl2l10 | -0.079 |
| cnga3a | 0.152 | cirbpb | -0.078 |
| rbp4l | 0.150 | vamp1 | -0.078 |
| ckmt2a | 0.149 | ndrg4 | -0.078 |
| gngt2b | 0.149 | stx3a | -0.076 |
| rcvrn3 | 0.148 | ppp1r1b | -0.075 |
| gucy2d | 0.148 | neurod4 | -0.074 |
| slc24a2 | 0.147 | scrt2 | -0.074 |
| rgs9a | 0.145 | nova2 | -0.074 |
| si:ch1073-469d17.2 | 0.144 | marcksl1a | -0.073 |
| rcvrn2 | 0.143 | btg1 | -0.071 |
| arr3a | 0.143 | lin7a | -0.071 |
| cngb3.2 | 0.140 | mpp6b | -0.071 |
| idh2 | 0.140 | dscaml1 | -0.069 |
| si:ch211-81a5.8 | 0.139 | nsg2 | -0.069 |
| aipl2 | 0.137 | bhlhe23 | -0.068 |
| opn1lw2 | 0.137 | gpm6aa | -0.067 |
| eno1b | 0.137 | zc4h2 | -0.067 |
| si:ch211-207l14.1 | 0.136 | gpm6ab | -0.066 |
| arl3l2 | 0.136 | gnao1b | -0.066 |
| tmem237a | 0.136 | hunk | -0.064 |
| inaa | 0.135 | ptmaa | -0.063 |
| hexb | 0.135 | epb41a | -0.063 |
| ppp1r18 | 0.133 | hmgn6 | -0.063 |