NK2 homeobox 2b
ZFIN 
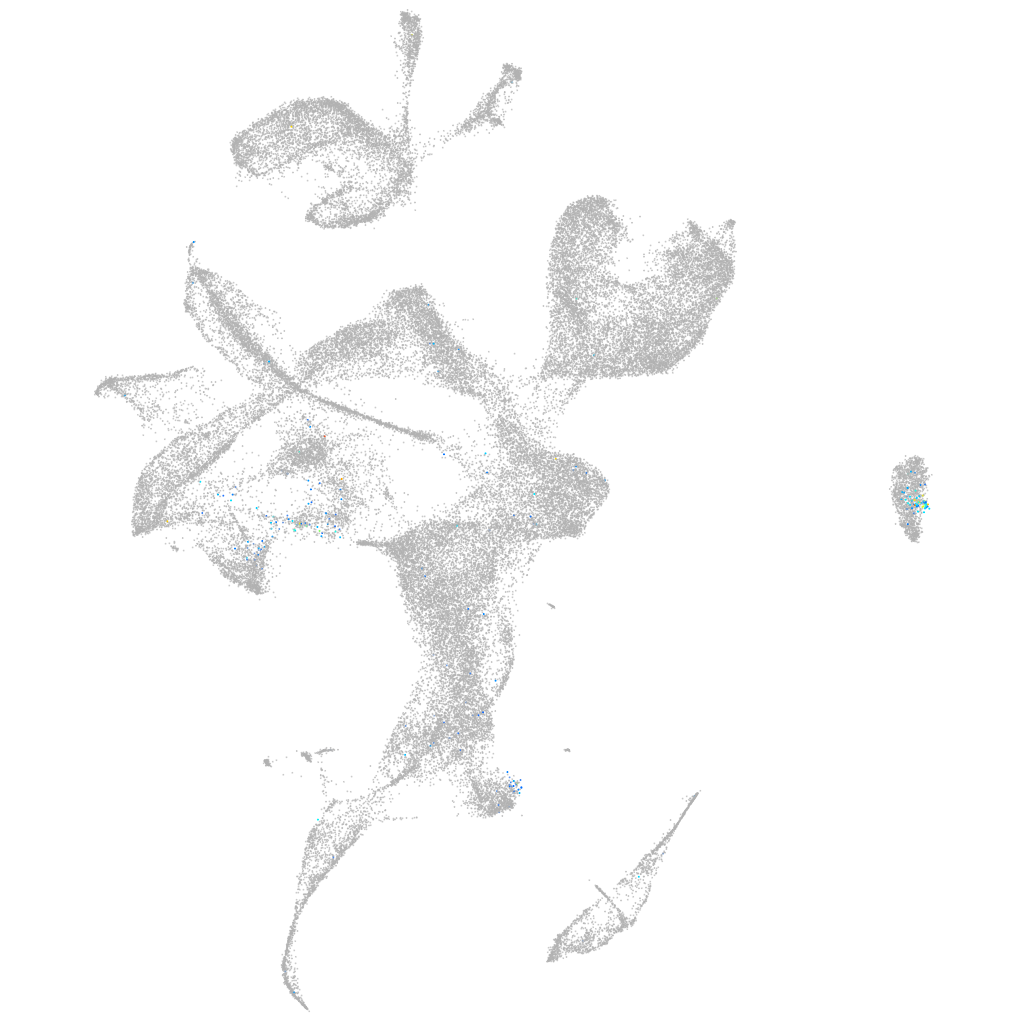
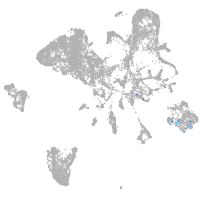
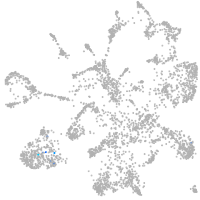
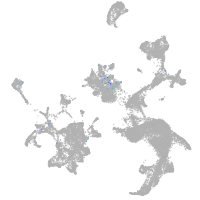
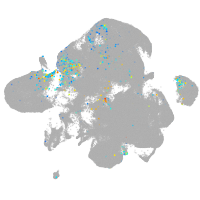
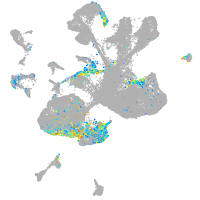
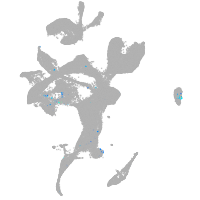
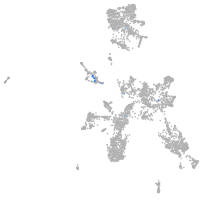
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lft1 | 0.348 | rps10 | -0.041 |
| nkx2.9 | 0.334 | rpl37 | -0.039 |
| nkx2.4b | 0.279 | ppiab | -0.036 |
| lft2 | 0.203 | zgc:158463 | -0.034 |
| ndr2 | 0.184 | zgc:114188 | -0.034 |
| gsc | 0.179 | ptmaa | -0.033 |
| foxb1b | 0.171 | COX3 | -0.032 |
| dact2 | 0.156 | h3f3a | -0.030 |
| nkx2.1 | 0.153 | CR383676.1 | -0.027 |
| dbx1a | 0.153 | mt-co2 | -0.027 |
| foxa | 0.150 | si:ch1073-429i10.3.1 | -0.026 |
| nkx2.4a | 0.148 | rorb | -0.026 |
| shha | 0.146 | cadm3 | -0.026 |
| si:dkey-34f9.3 | 0.145 | pvalb1 | -0.026 |
| nkx1.2la | 0.145 | hnrnpa0l | -0.025 |
| LOC103909868 | 0.135 | atp5mc1 | -0.025 |
| sox19a | 0.135 | ckbb | -0.025 |
| foxd5 | 0.130 | rps17 | -0.025 |
| LOC100331751 | 0.129 | gpm6ab | -0.024 |
| nkx2.2a | 0.123 | zgc:153409 | -0.024 |
| LOC101883163 | 0.109 | pvalb2 | -0.023 |
| tdgf1 | 0.105 | foxg1b | -0.023 |
| hspb1 | 0.103 | actc1b | -0.023 |
| fzd8b | 0.103 | atp6v1e1b | -0.022 |
| foxa3 | 0.103 | hbae3 | -0.022 |
| ptch2 | 0.102 | hbbe1.3 | -0.022 |
| CR847803.1 | 0.101 | cox5aa | -0.022 |
| NAV1 | 0.100 | snap25b | -0.022 |
| BX088596.2 | 0.100 | gapdhs | -0.022 |
| fgf3 | 0.098 | pax6a | -0.022 |
| shisa2a | 0.097 | mt-nd1 | -0.022 |
| BX005308.1 | 0.097 | atp5f1b | -0.021 |
| epha4a | 0.095 | syt5b | -0.021 |
| sox1a | 0.093 | mt-atp6 | -0.021 |
| rfx4 | 0.092 | gpm6aa | -0.021 |