sodium/potassium transporting ATPase interacting 4
ZFIN 
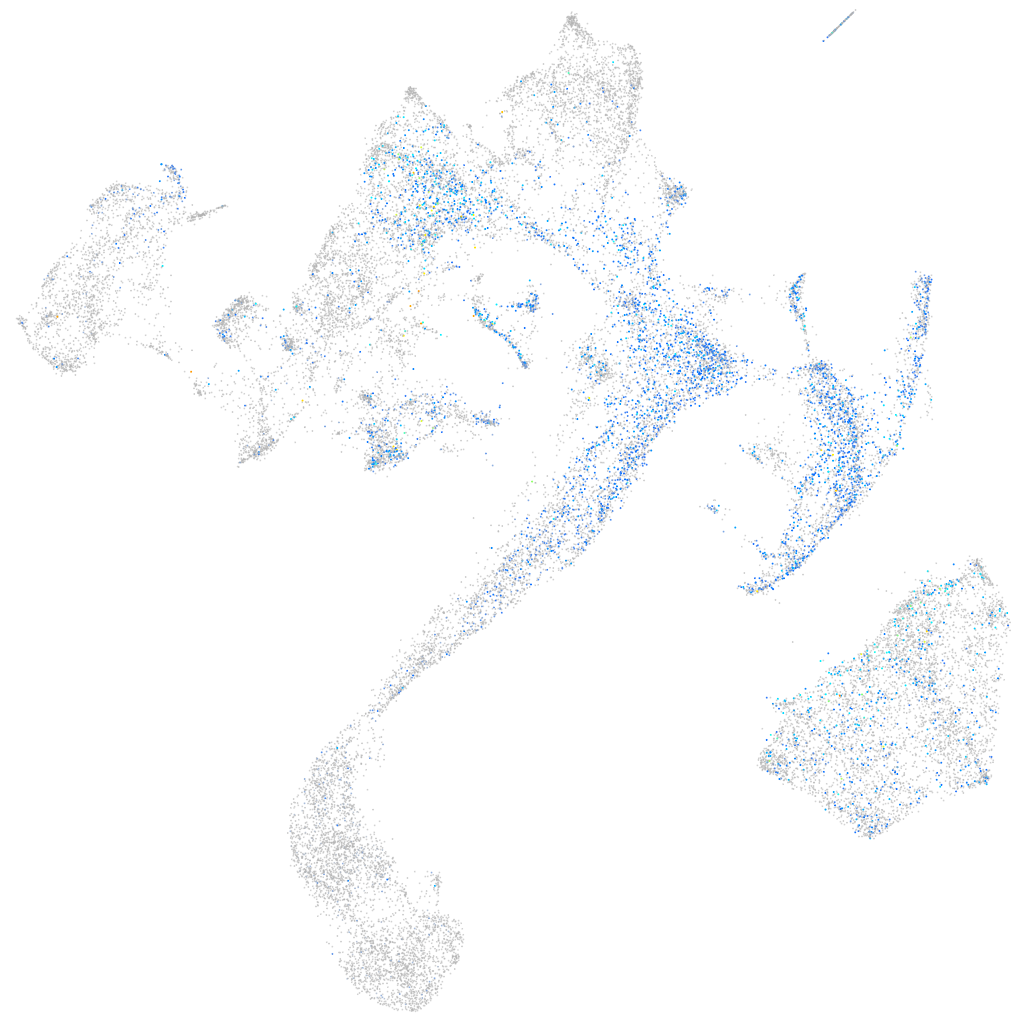
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fn1b | 0.215 | ckma | -0.172 |
| cdh2 | 0.197 | ckmb | -0.169 |
| hsp90ab1 | 0.179 | atp2a1 | -0.164 |
| naca | 0.169 | actc1b | -0.161 |
| ran | 0.167 | ak1 | -0.161 |
| eef1a1l1 | 0.166 | gapdh | -0.160 |
| ubc | 0.164 | tnnc2 | -0.154 |
| mdka | 0.163 | si:ch73-367p23.2 | -0.152 |
| h3f3d | 0.159 | mylpfa | -0.152 |
| XLOC-001964 | 0.154 | neb | -0.151 |
| eef2b | 0.153 | tnnt3a | -0.151 |
| cirbpb | 0.152 | mylz3 | -0.150 |
| id1 | 0.151 | ldb3b | -0.149 |
| h2afvb | 0.151 | pvalb1 | -0.148 |
| tcf15 | 0.150 | aldoab | -0.146 |
| BX001014.2 | 0.148 | actn3a | -0.146 |
| tspan7 | 0.145 | ank1a | -0.146 |
| khdrbs1a | 0.144 | eno3 | -0.144 |
| snrpf | 0.144 | pvalb2 | -0.144 |
| hnrnpa0l | 0.143 | myom1a | -0.144 |
| pabpc1a | 0.143 | smyd1a | -0.143 |
| nop53 | 0.142 | pgam2 | -0.143 |
| srsf5a | 0.141 | tmem38a | -0.142 |
| rsl24d1 | 0.141 | acta1b | -0.141 |
| si:ch73-281n10.2 | 0.140 | prx | -0.140 |
| eif3ba | 0.140 | tmod4 | -0.140 |
| hmga1a | 0.139 | tnnt3b | -0.139 |
| meox1 | 0.138 | si:ch211-266g18.10 | -0.139 |
| eif3m | 0.138 | nme2b.2 | -0.138 |
| eif3f | 0.137 | hhatla | -0.137 |
| hnrnpaba | 0.137 | tnni2a.4 | -0.137 |
| syncrip | 0.137 | XLOC-025819 | -0.137 |
| eif3k | 0.137 | CABZ01078594.1 | -0.136 |
| cbx3a | 0.136 | cav3 | -0.135 |
| hspa8 | 0.136 | si:dkey-16p21.8 | -0.135 |