nipsnap homolog 2
ZFIN 
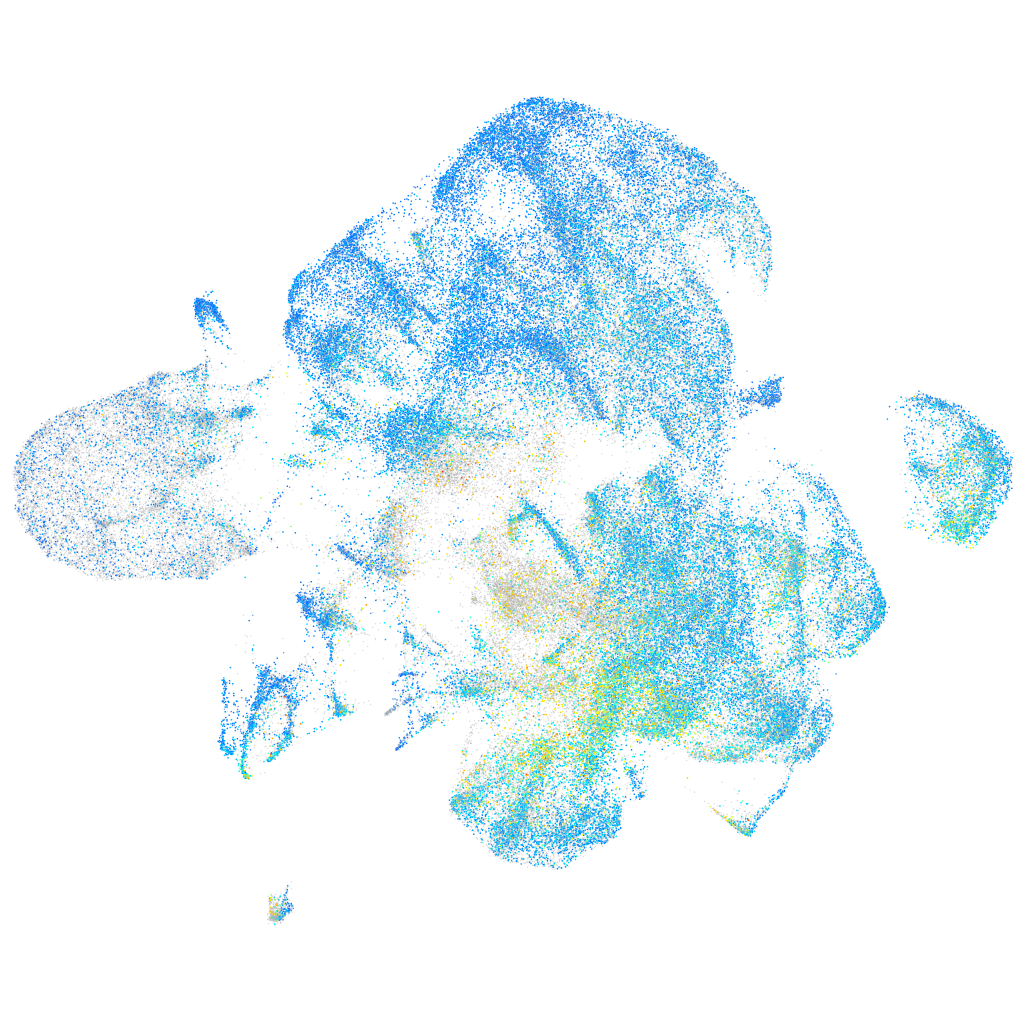
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.202 | NC-002333.4 | -0.143 |
| tpi1b | 0.197 | si:ch211-152c2.3 | -0.127 |
| atp5mc1 | 0.196 | stm | -0.120 |
| atp6v1g1 | 0.193 | apoeb | -0.115 |
| atp6v1e1b | 0.188 | apoc1 | -0.114 |
| ppiab | 0.187 | crabp2b | -0.107 |
| calm3b | 0.182 | pou5f3 | -0.106 |
| ldhba | 0.181 | hspb1 | -0.105 |
| atp5f1b | 0.179 | akap12b | -0.100 |
| vdac1 | 0.178 | polr3gla | -0.099 |
| ywhag2 | 0.176 | COX7A2 | -0.097 |
| cox4i1 | 0.172 | hmgb2a | -0.096 |
| eno1a | 0.170 | zmp:0000000624 | -0.096 |
| calm1a | 0.169 | si:dkey-66i24.9 | -0.096 |
| zgc:65894 | 0.169 | s100a1 | -0.093 |
| ndufa4 | 0.168 | hmga1a | -0.089 |
| COX5B | 0.168 | alcamb | -0.088 |
| calm1b | 0.166 | vox | -0.087 |
| atp5mc3b | 0.166 | arf1 | -0.087 |
| gng3 | 0.166 | ved | -0.086 |
| si:ch1073-429i10.3.1 | 0.166 | zgc:110425 | -0.086 |
| syn2a | 0.166 | shisa2a | -0.086 |
| snap25a | 0.164 | nr6a1a | -0.085 |
| cox7a2a | 0.163 | pkdccb | -0.085 |
| vdac3 | 0.163 | sp5l | -0.085 |
| ywhabl | 0.163 | asb11 | -0.085 |
| cox8a | 0.163 | LOC108190024 | -0.085 |
| stmn2a | 0.162 | si:ch1073-80i24.3 | -0.084 |
| calm2a | 0.162 | apela | -0.082 |
| atp5meb | 0.162 | si:dkey-92i17.2 | -0.080 |
| sncgb | 0.161 | ISCU | -0.079 |
| sncb | 0.160 | cdx4 | -0.079 |
| tuba1c | 0.160 | irx7 | -0.078 |
| necap1 | 0.159 | mki67 | -0.078 |
| atp5if1b | 0.158 | nnr | -0.077 |